The ubiquitous hammerhead ribozyme
- PMID: 22454536
- PMCID: PMC3334697
- DOI: 10.1261/rna.031401.111
The ubiquitous hammerhead ribozyme
Abstract
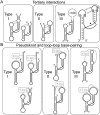
The hammerhead ribozyme is a small catalytic RNA motif capable of endonucleolytic (self-) cleavage. It is composed of a catalytic core of conserved nucleotides flanked by three helices, two of which form essential tertiary interactions for fast self-scission under physiological conditions. Originally discovered in subviral plant pathogens, its presence in several eukaryotic genomes has been reported since. More recently, this catalytic RNA motif has been shown to reside in a large number of genomes. We review the different approaches in discovering these new hammerhead ribozyme sequences and discuss possible biological functions of the genomic motifs.
Figures




References
-
- Branch AD, Robertson HD 1984. A replication cycle for viroids and other small infectious RNAs. Science 223: 450–455 - PubMed
-
- Buzayan JM, Gerlach WL, Bruening G 1986. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature 323: 349–353
-
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A 2004. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc 126: 10848–10849 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
