Epigenetic features are significantly associated with alternative splicing
- PMID: 22455468
- PMCID: PMC3362759
- DOI: 10.1186/1471-2164-13-123
Epigenetic features are significantly associated with alternative splicing
Abstract
Background: While alternative splicing (AS) contributes greatly to protein diversities, the relationship between various types of AS and epigenetic factors remains largely unknown.
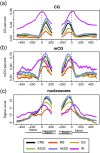
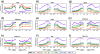
Results: In this study, we discover that a number of epigenetic features, including DNA methylation, nucleosome occupancy, specific histone modifications and protein features, are strongly associated with AS. To further enhance our understanding of the association between these features and AS, we cluster our investigated features based on their association patterns with each AS type into four groups, with H3K36me3, EGR1, GABP, SRF, SIN3A and RNA Pol II grouped together and showing strongest association with AS. In addition, we find that the AS types can be classified into two general classes, namely the exon skipping related process (ESRP), and the alternative splice site selection process (ASSP), based on their association levels with the epigenetic features.
Conclusion: Our analysis thus suggests that epigenetic features are likely to play important roles in regulating AS.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

