Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes
- PMID: 22455878
- PMCID: PMC3439974
- DOI: 10.1186/gb-2012-13-3-r23
Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes
Abstract
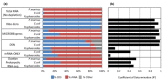
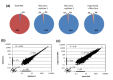
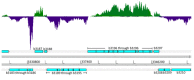
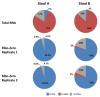
We have developed a process for transcriptome analysis of bacterial communities that accommodates both intact and fragmented starting RNA and combines efficient rRNA removal with strand-specific RNA-seq. We applied this approach to an RNA mixture derived from three diverse cultured bacterial species and to RNA isolated from clinical stool samples. The resulting expression profiles were highly reproducible, enriched up to 40-fold for non-rRNA transcripts, and correlated well with profiles representing undepleted total RNA.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

