Coprolites as a source of information on the genome and diet of the cave hyena
- PMID: 22456883
- PMCID: PMC3367792
- DOI: 10.1098/rspb.2012.0358
Coprolites as a source of information on the genome and diet of the cave hyena
Abstract
We performed high-throughput sequencing of DNA from fossilized faeces to evaluate this material as a source of information on the genome and diet of Pleistocene carnivores. We analysed coprolites derived from the extinct cave hyena (Crocuta crocuta spelaea), and sequenced 90 million DNA fragments from two specimens. The DNA reads enabled a reconstruction of the cave hyena mitochondrial genome with up to a 158-fold coverage. This genome, and those sequenced from extant spotted (Crocuta crocuta) and striped (Hyaena hyaena) hyena specimens, allows for the establishment of a robust phylogeny that supports a close relationship between the cave and the spotted hyena. We also demonstrate that high-throughput sequencing yields data for cave hyena multi-copy and single-copy nuclear genes, and that about 50 per cent of the coprolite DNA can be ascribed to this species. Analysing the data for additional species to indicate the cave hyena diet, we retrieved abundant sequences for the red deer (Cervus elaphus), and characterized its mitochondrial genome with up to a 3.8-fold coverage. In conclusion, we have demonstrated the presence of abundant ancient DNA in the coprolites surveyed. Shotgun sequencing of this material yielded a wealth of DNA sequences for a Pleistocene carnivore and allowed unbiased identification of diet.
Figures

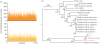


References
-
- Higuchi R., Bowman B., Freiberger M., Ryder O. A., Wilson A. C. 1984. DNA sequences from the quagga, an extinct member of the horse family. Nature 312, 282–284 10.1038/312282a0 (doi:10.1038/312282a0) - DOI - PubMed
-
- Cooper A., Lalueza-Fox C., Anderson S., Rambaut A., Austin J., Ward R. 2001. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 409, 704–707 10.1038/35055536 (doi:10.1038/35055536) - DOI - PubMed
-
- Haddrath O., Baker A. J. 2001. Complete mitochondrial DNA genome sequences of extinct birds: ratite phylogenetics and the vicariance biogeography hypothesis. Proc. R. Soc. Lond. B 268, 939–945 10.1098/rspb.2001.1587 (doi:10.1098/rspb.2001.1587) - DOI - PMC - PubMed
-
- Poinar H. N., et al. 2006. Metagenomics to paleogenomics, large-scale sequencing of mammoth DNA. Science 311, 392–394 10.1126/science.1123360 (doi:10.1126/science.1123360) - DOI - PubMed
-
- Miller W., et al. 2008 Sequencing the nuclear genome of the extinct woolly mammoth. Nature 456, 387–390 10.1038/nature07446 (doi:10.1038/nature07446) - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
