Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding
- PMID: 22457068
- PMCID: PMC3401448
- DOI: 10.1093/nar/gks263
Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding
Abstract
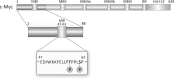
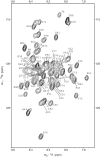
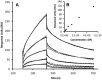
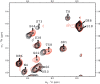
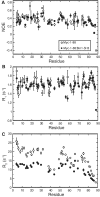
The crucial role of Myc as an oncoprotein and as a key regulator of cell growth makes it essential to understand the molecular basis of Myc function. The N-terminal region of c-Myc coordinates a wealth of protein interactions involved in transformation, differentiation and apoptosis. We have characterized in detail the intrinsically disordered properties of Myc-1-88, where hierarchical phosphorylation of S62 and T58 regulates activation and destruction of the Myc protein. By nuclear magnetic resonance (NMR) chemical shift analysis, relaxation measurements and NOE analysis, we show that although Myc occupies a very heterogeneous conformational space, we find transiently structured regions in residues 22-33 and in the Myc homology box I (MBI; residues 45-65); both these regions are conserved in other members of the Myc family. Binding of Bin1 to Myc-1-88 as assayed by NMR and surface plasmon resonance (SPR) revealed primary binding to the S62 region in a dynamically disordered and multivalent complex, accompanied by population shifts leading to altered intramolecular conformational dynamics. These findings expand the increasingly recognized concept of intrinsically disordered regions mediating transient interactions to Myc, a key transcriptional regulator of major medical importance, and have important implications for further understanding its multifaceted role in gene regulation.
Figures