A mostly traditional approach improves alignment of bisulfite-converted DNA
- PMID: 22457070
- PMCID: PMC3401460
- DOI: 10.1093/nar/gks275
A mostly traditional approach improves alignment of bisulfite-converted DNA
Abstract
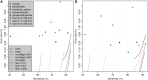
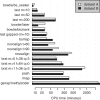
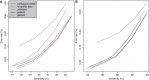
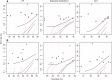
Cytosines in genomic DNA are sometimes methylated. This affects many biological processes and diseases. The standard way of measuring methylation is to use bisulfite, which converts unmethylated cytosines to thymines, then sequence the DNA and compare it to a reference genome sequence. We describe a method for the critical step of aligning the DNA reads to the correct genomic locations. Our method builds on classic alignment techniques, including likelihood-ratio scores and spaced seeds. In a realistic benchmark, our method has a better combination of sensitivity, specificity and speed than nine other high-throughput bisulfite aligners. This study enables more accurate and rational analysis of DNA methylation. It also illustrates how to adapt general-purpose alignment methods to a special case with distorted base patterns: this should be informative for other special cases such as ancient DNA and AT-rich genomes.
Figures

References
-
- Watanabe Y, Maekawa M. Methylation of DNA in cancer. Adv. Clin. Chem. 2010;52:145–167. - PubMed
-
- Vanyushin BF, Ashapkin VV. DNA methylation in higher plants: past, present and future. Biochim. Biophys. Acta. 2011;1809:360–368. - PubMed
-
- Coppieters N, Dragunow M. Epigenetics in Alzheimer's disease: a focus on DNA modifications. Curr. Pharm. Des. 2011;17:3398–3412. - PubMed
-
- Mund C, Lyko F. Epigenetic cancer therapy: Proof of concept and remaining challenges. Bioessays. 2010;32:949–957. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

