Mechanisms for defining supercoiling set point of DNA gyrase orthologs: II. The shape of the GyrA subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency
- PMID: 22457352
- PMCID: PMC3365746
- DOI: 10.1074/jbc.M112.345736
Mechanisms for defining supercoiling set point of DNA gyrase orthologs: II. The shape of the GyrA subunit C-terminal domain (CTD) is not a sole determinant for controlling supercoiling efficiency
Abstract
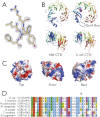
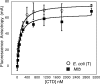
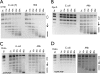
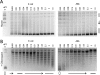
DNA topoisomerases are essential enzymes that can overwind, underwind, and disentangle double-helical DNA segments to maintain the topological state of chromosomes. Nearly all bacteria utilize a unique type II topoisomerase, gyrase, which actively adds negative supercoils to chromosomes using an ATP-dependent DNA strand passage mechanism; however, the specific activities of these enzymes can vary markedly from species to species. Escherichia coli gyrase is known to favor supercoiling over decatenation (Zechiedrich, E. L., Khodursky, A. B., and Cozzarelli, N. R. (1997) Genes Dev. 11, 2580-2592), whereas the opposite has been reported for Mycobacterium tuberculosis gyrase (Aubry, A., Fisher, L. M., Jarlier, V., and Cambau, E. (2006) Biochem. Biophys. Res. Commun. 348, 158-165). Here, we set out to understand the molecular basis for these differences using structural and biochemical approaches. Contrary to expectations based on phylogenetic inferences, we find that the dedicated DNA wrapping domains (the C-terminal domains) of both gyrases are highly similar, both architecturally and in their ability to introduce writhe into DNA. However, the M. tuberculosis enzyme lacks a C-terminal control element recently uncovered in E. coli gyrase (see accompanying article (Tretter, E. M., and Berger, J. M. (2012) J. Biol. Chem. 287, 18636-18644)) and turns over ATP at a much slower rate. Together, these findings demonstrate that C-terminal domain shape is not the sole regulatory determinant of gyrase activity and instead indicate that an inability to tightly couple DNA wrapping to ATP turnover is why M. tuberculosis gyrase cannot supercoil DNA to the same extent as its γ-proteobacterial counterpart. Our observations demonstrate that gyrase has been modified in multiple ways throughout evolution to fine-tune its specific catalytic properties.
Figures



References
-
- Zechiedrich E. L., Khodursky A. B., Bachellier S., Schneider R., Chen D., Lilley D. M., Cozzarelli N. R. (2000) Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 275, 8103–8113 - PubMed
-
- Drlica K. (1990) Bacterial topoisomerases and the control of DNA supercoiling. Trends Genet. 6, 433–437 - PubMed
-
- Forterre P., Gribaldo S., Gadelle D., Serre M. C. (2007) Origin and evolution of DNA topoisomerases. Biochimie 89, 427–446 - PubMed
-
- Booker B. M., Deng S., Higgins N. P. (2010) DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 78, 1348–1364 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

