Metabolic flux increases glycoprotein sialylation: implications for cell adhesion and cancer metastasis
- PMID: 22457533
- PMCID: PMC3394959
- DOI: 10.1074/mcp.M112.017558
Metabolic flux increases glycoprotein sialylation: implications for cell adhesion and cancer metastasis
Abstract
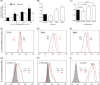
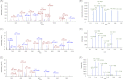
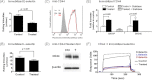
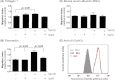
This study reports a global glycoproteomic analysis of pancreatic cancer cells that describes how flux through the sialic acid biosynthetic pathway selectively modulates a subset of N-glycosylation sites found within cellular proteins. These results provide evidence that sialoglycoprotein patterns are not determined exclusively by the transcription of biosynthetic enzymes or the availability of N-glycan sequons; instead, bulk metabolic flux through the sialic acid pathway has a remarkable ability to increase the abundance of certain sialoglycoproteins while having a minimal impact on others. Specifically, of 82 glycoproteins identified through a mass spectrometry and bioinformatics approach, ≈ 31% showed no change in sialylation, ≈ 29% exhibited a modest increase, whereas ≈ 40% experienced an increase of greater than twofold. Increased sialylation of specific glycoproteins resulted in changes to the adhesive properties of SW1990 pancreatic cancer cells (e.g. increased CD44-mediated adhesion to selectins under physiological flow and enhanced integrin-mediated cell mobility on collagen and fibronectin). These results indicate that cancer cells can become more aggressively malignant by controlling the sialylation of proteins implicated in metastatic transformation via metabolic flux.
Conflict of interest statement
Figures



References
-
- Monica T. J., Andersen D. C., Goochee C. F. (1997) A mathematical model of sialylation of N-linked oligosaccharides in the trans-Golgi network. Glycobiology 7, 515–521 - PubMed
-
- Lau K. S., Partridge E. A., Grigorian A., Silvescu C. I., Reinhold V. N., Demetriou M., Dennis J. W. (2007) Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 - PubMed
-
- Lau K. S., Dennis J. W. (2008) N-Glycans in cancer progression. Glycobiology 18, 750–760 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

