Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease
- PMID: 22457623
- PMCID: PMC3310798
- DOI: 10.1371/journal.ppat.1002601
Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease
Abstract
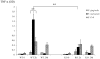
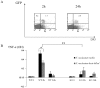
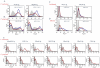
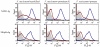
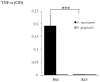
Periodontitis is a common human chronic inflammatory disease that results in the destruction of the tooth attachment apparatus and tooth loss. Although infections with periopathogenic bacteria such as Porphyromonas gingivalis (P. gingivalis) and Fusobacterium nucleatum (F. nucleatum) are essential for inducing periodontitis, the nature and magnitude of the disease is determined by the host's immune response. Here, we investigate the role played by the NK killer receptor NKp46 (NCR1 in mice), in the pathogenesis of periodontitis. Using an oral infection periodontitis model we demonstrate that following F. nucleatum infection no alveolar bone loss is observed in mice deficient for NCR1 expression, whereas around 20% bone loss is observed in wild type mice and in mice infected with P. gingivalis. By using subcutaneous chambers inoculated with F. nucleatum we demonstrate that immune cells, including NK cells, rapidly accumulate in the chambers and that this leads to a fast and transient, NCR1-dependant TNF-α secretion. We further show that both the mouse NCR1 and the human NKp46 bind directly to F. nucleatum and we demonstrate that this binding is sensitive to heat, to proteinase K and to pronase treatments. Finally, we show in vitro that the interaction of NK cells with F. nucleatum leads to an NCR1-dependent secretion of TNF-α. Thus, the present study provides the first evidence that NCR1 and NKp46 directly recognize a periodontal pathogen and that this interaction influences the outcome of F. nucleatum-mediated periodontitis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Albandar JM, Brunelle JA, Kingman A. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988–1994. J Periodontol. 1999;70:13–29. - PubMed
-
- Wu T, Trevisan M, Genco RJ, Dorn JP, Falkner KL, et al. Periodontal disease and risk of cerebrovascular disease: the first national health and nutrition examination survey and its follow-up study. Arch Intern Med. 2000;160:2749–2755. - PubMed
-
- Scannapieco FA. Role of oral bacteria in respiratory infection. J Periodontol. 1999;70:793–802. - PubMed
-
- Offenbacher S, Jared HL, O'Reilly PG, Wells SR, Salvi GE, et al. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann Periodontol. 1998;3:233–250. - PubMed
-
- Dodman T, Robson J, Pincus D. Kingella kingae infections in children. J Paediatr Child Health. 2000;36:87–90. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

