Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity
- PMID: 22457629
- PMCID: PMC3310836
- DOI: 10.1371/journal.ppat.1002612
Thriving under stress: selective translation of HIV-1 structural protein mRNA during Vpr-mediated impairment of eIF4E translation activity
Abstract
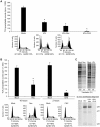
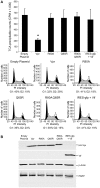
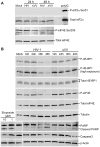
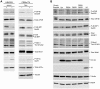
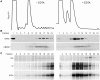
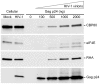
Translation is a regulated process and is pivotal to proper cell growth and homeostasis. All retroviruses rely on the host translational machinery for viral protein synthesis and thus may be susceptible to its perturbation in response to stress, co-infection, and/or cell cycle arrest. HIV-1 infection arrests the cell cycle in the G2/M phase, potentially disrupting the regulation of host cell translation. In this study, we present evidence that HIV-1 infection downregulates translation in lymphocytes, attributable to the cell cycle arrest induced by the HIV-1 accessory protein Vpr. The molecular basis of the translation suppression is reduced accumulation of the active form of the translation initiation factor 4E (eIF4E). However, synthesis of viral structural proteins is sustained despite the general suppression of protein production. HIV-1 mRNA translation is sustained due to the distinct composition of the HIV-1 ribonucleoprotein complexes. RNA-coimmunoprecipitation assays determined that the HIV-1 unspliced and singly spliced transcripts are predominantly associated with nuclear cap binding protein 80 (CBP80) in contrast to completely-spliced viral and cellular mRNAs that are associated with eIF4E. The active translation of the nuclear cap binding complex (CBC)-bound viral mRNAs is demonstrated by ribosomal RNA profile analyses. Thus, our findings have uncovered that the maintenance of CBC association is a novel mechanism used by HIV-1 to bypass downregulation of eIF4E activity and sustain viral protein synthesis. We speculate that a subset of CBP80-bound cellular mRNAs contribute to recovery from significant cellular stress, including human retrovirus infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

