APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete "all or nothing" phenomenon
- PMID: 22457633
- PMCID: PMC3310730
- DOI: 10.1371/journal.pgen.1002550
APOBEC3G-induced hypermutation of human immunodeficiency virus type-1 is typically a discrete "all or nothing" phenomenon
Abstract
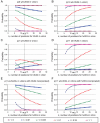
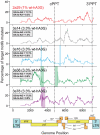
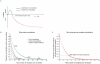
The rapid evolution of Human Immunodeficiency Virus (HIV-1) allows studies of ongoing host-pathogen interactions. One key selective host factor is APOBEC3G (hA3G) that can cause extensive and inactivating Guanosine-to-Adenosine (G-to-A) mutation on HIV plus-strand DNA (termed hypermutation). HIV can inhibit this innate anti-viral defense through binding of the viral protein Vif to hA3G, but binding efficiency varies and hypermutation frequencies fluctuate in patients. A pivotal question is whether hA3G-induced G-to-A mutation is always lethal to the virus or if it may occur at sub-lethal frequencies that could increase viral diversification. We show in vitro that limiting-levels of hA3G-activity (i.e. when only a single hA3G-unit is likely to act on HIV) produce hypermutation frequencies similar to those in patients and demonstrate in silico that potentially non-lethal G-to-A mutation rates are ∼10-fold lower than the lowest observed hypermutation levels in vitro and in vivo. Our results suggest that even a single incorporated hA3G-unit is likely to cause extensive and inactivating levels of HIV hypermutation and that hypermutation therefore is typically a discrete "all or nothing" phenomenon. Thus, therapeutic measures that inhibit the interaction between Vif and hA3G will likely not increase virus diversification but expand the fraction of hypermutated proviruses within the infected host.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. - PubMed
-
- Hache G, Mansky LM, Harris RS. Human APOBEC3 proteins, retrovirus restriction, and HIV drug resistance. AIDS Rev. 2006;8:148–57. - PubMed
-
- Simon V, Zennou V, Murray D, Huang Y, Ho DD, et al. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

