Archaeal origin of tubulin
- PMID: 22458654
- PMCID: PMC3349469
- DOI: 10.1186/1745-6150-7-10
Archaeal origin of tubulin
Abstract
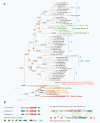
Tubulins are a family of GTPases that are key components of the cytoskeleton in all eukaryotes and are distantly related to the FtsZ GTPase that is involved in cell division in most bacteria and many archaea. Among prokaryotes, bona fide tubulins have been identified only in bacteria of the genus Prosthecobacter. These bacterial tubulin genes appear to have been horizontally transferred from eukaryotes. Here we describe tubulins encoded in the genomes of thaumarchaeota of the genus Nitrosoarchaeum that we denote artubulins Phylogenetic analysis results are compatible with the origin of eukaryotic tubulins from artubulins. These findings expand the emerging picture of the origin of key components of eukaryotic functional systems from ancestral forms that are scattered among the extant archaea.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

