A comparative proteomic study identified LRPPRC and MCM7 as putative actors in imatinib mesylate cross-resistance in Lucena cell line
- PMID: 22458888
- PMCID: PMC3361502
- DOI: 10.1186/1477-5956-10-23
A comparative proteomic study identified LRPPRC and MCM7 as putative actors in imatinib mesylate cross-resistance in Lucena cell line
Abstract
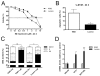
Background: Although chronic myeloid leukemia (CML) treatment has improved since the introduction of imatinib mesylate (IM), cases of resistance have been reported. This resistance has been associated with the emergence of multidrug resistance (MDR) phenotype, as a BCR-ABL independent mechanism. The classic pathway studied in MDR promotion is ATP-binding cassette (ABC) family transporters expression, but other mechanisms that drive drug resistance are largely unknown. To better understand IM therapy relapse due to the rise of MDR, we compared the proteomic profiles of K562 and Lucena (K562/VCR) cells.
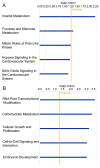
Results: The use of 2-DE coupled with a MS approach resulted in the identification of 36 differentially expressed proteins. Differential mRNA levels of leucine-rich PPR motif-containing (LRPPRC) protein, minichromosome maintenance complex component 7 (MCM7) and ATP-binding cassette sub-family B (MDR/TAP) member 1 (ABCB1) were capable of defining samples from CML patients as responsive or resistant to therapy.
Conclusions: Through the data presented in this work, we show the relevance of MDR to IM therapy. In addition, our proteomic approach identified candidate actors involved in resistance, which could lead to additional information on BCR-ABL-independent molecular mechanisms.
Figures







References
-
- Jiang X, Zhao Y, Smith C, Gasparetto M, Turhan A, Eaves A, Eaves C. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21:926–935. - PubMed
-
- Quintás-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of Primary and Secondary Resistance to Imatinib in Chronic Myeloid Leukemia. Cancer Control. 2009;16:122–131. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

