Sindbis virus induces the production of a novel class of endogenous siRNAs in Aedes aegypti mosquitoes
- PMID: 22458920
- PMCID: PMC3386798
- DOI: 10.1111/j.1365-2583.2012.01141.x
Sindbis virus induces the production of a novel class of endogenous siRNAs in Aedes aegypti mosquitoes
Abstract
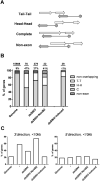
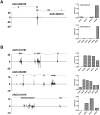
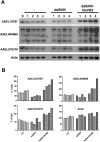
Small RNA regulatory pathways are used to control the activity of transposons, regulate gene expression and resist infecting viruses. We examined the biogenesis of mRNA-derived endogenous short-interfering RNAs (endo-siRNAs) in the disease vector mosquito Aedes aegypti. Under standard conditions, mRNA-derived endo-siRNAs were produced from the bidirectional transcription of tail-tail overlapping gene pairs. Upon infection with the alphavirus, Sindbis virus (SINV), another class of mRNA-derived endo-siRNAs was observed. Genes producing SINV-induced endo-siRNAs were not enriched for overlapping partners or nearby genes, but were enriched for transcripts with long 3' untranslated regions. Endo-siRNAs from this class derived uniformly from the entire length of the target transcript, and were found to regulate the transcript levels of the genes from which they were derived. Strand-specific quantitative PCR experiments demonstrated that antisense strands of targeted mRNA genes were produced to exonic, but not intronic regions. Finally, small RNAs mapped to both sense and antisense strands of exon-exon junctions, suggesting double-stranded RNA precursors to SINV-induced endo-siRNAs may be synthesized from mature mRNA templates. These results suggest additional complexity in small RNA pathways and gene regulation in the presence of an infecting virus in disease vector mosquitoes.
© 2012 The Authors. Insect Molecular Biology © 2012 The Royal Entomological Society.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

