TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements
- PMID: 22460799
- PMCID: PMC3341064
- DOI: 10.1073/pnas.1203028109
TLS/FUS (translocated in liposarcoma/fused in sarcoma) regulates target gene transcription via single-stranded DNA response elements
Abstract
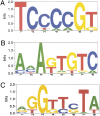
TLS/FUS (TLS) is a multifunctional protein implicated in a wide range of cellular processes, including transcription and mRNA processing, as well as in both cancer and neurological disease. However, little is currently known about TLS target genes and how they are recognized. Here, we used ChIP and promoter microarrays to identify genes potentially regulated by TLS. Among these genes, we detected a number that correlate with previously known functions of TLS, and confirmed TLS occupancy at several of them by ChIP. We also detected changes in mRNA levels of these target genes in cells where TLS levels were altered, indicative of both activation and repression. Next, we used data from the microarray and computational methods to determine whether specific sequences were enriched in DNA fragments bound by TLS. This analysis suggested the existence of TLS response elements, and we show that purified TLS indeed binds these sequences with specificity in vitro. Remarkably, however, TLS binds only single-strand versions of the sequences. Taken together, our results indicate that TLS regulates expression of specific target genes, likely via recognition of specific single-stranded DNA sequences located within their promoter regions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
-
- Crozat A, Aman P, Mandahl N, Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. - PubMed
-
- Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

