Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β
- PMID: 22460952
- PMCID: PMC3367514
- DOI: 10.1038/nature11048
Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β
Abstract
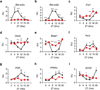
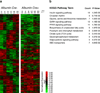
The circadian clock acts at the genomic level to coordinate internal behavioural and physiological rhythms via the CLOCK-BMAL1 transcriptional heterodimer. Although the nuclear receptors REV-ERB-α and REV-ERB-β have been proposed to form an accessory feedback loop that contributes to clock function, their precise roles and importance remain unresolved. To establish their regulatory potential, we determined the genome-wide cis-acting targets (cistromes) of both REV-ERB isoforms in murine liver, which revealed shared recognition at over 50% of their total DNA binding sites and extensive overlap with the master circadian regulator BMAL1. Although REV-ERB-α has been shown to regulate Bmal1 expression directly, our cistromic analysis reveals a more profound connection between BMAL1 and the REV-ERB-α and REV-ERB-β genomic regulatory circuits than was previously suspected. Genes within the intersection of the BMAL1, REV-ERB-α and REV-ERB-β cistromes are highly enriched for both clock and metabolic functions. As predicted by the cistromic analysis, dual depletion of Rev-erb-α and Rev-erb-β function by creating double-knockout mice profoundly disrupted circadian expression of core circadian clock and lipid homeostatic gene networks. As a result, double-knockout mice show markedly altered circadian wheel-running behaviour and deregulated lipid metabolism. These data now unite REV-ERB-α and REV-ERB-β with PER, CRY and other components of the principal feedback loop that drives circadian expression and indicate a more integral mechanism for the coordination of circadian rhythm and metabolism.
Figures


Comment in
-
Drug discovery: Time in a bottle.Nature. 2012 May 2;485(7396):45-6. doi: 10.1038/485045a. Nature. 2012. PMID: 22552089 No abstract available.
-
REV-ERBs: more than the sum of the individual parts.Cell Metab. 2012 Jun 6;15(6):791-3. doi: 10.1016/j.cmet.2012.05.006. Cell Metab. 2012. PMID: 22682217
References
-
- Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. - PubMed
-
- Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. - PubMed
-
- Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 HL105278/HL/NHLBI NIH HHS/United States
- T32 HL007770/HL/NHLBI NIH HHS/United States
- DK091618/DK/NIDDK NIH HHS/United States
- DK062434/DK/NIDDK NIH HHS/United States
- U19 DK062434/DK/NIDDK NIH HHS/United States
- DK090962/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32-HL007770/HL/NHLBI NIH HHS/United States
- HL105278/HL/NHLBI NIH HHS/United States
- DK057978/DK/NIDDK NIH HHS/United States
- R24 DK090962/DK/NIDDK NIH HHS/United States
- R37 DK057978/DK/NIDDK NIH HHS/United States
- R01 DK091618/DK/NIDDK NIH HHS/United States
- R01 DK057978/DK/NIDDK NIH HHS/United States
- P30 CA014195/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

