Functional parameters of Dicer-independent microRNA biogenesis
- PMID: 22461413
- PMCID: PMC3334703
- DOI: 10.1261/rna.032938.112
Functional parameters of Dicer-independent microRNA biogenesis
Abstract
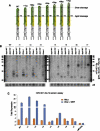
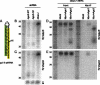
Until recently, a Dicer-class RNase III enzyme was believed to be essential for microRNA (miRNA) biogenesis in all animals. The conserved vertebrate locus mir-451 defies this expectation and instead matures by direct cleavage of its pre-miRNA hairpin via the Slicer activity of Argonaute2 (Ago2). In this study, we used structure-function analysis to define the functional parameters of Ago2-mediated miRNA biogenesis. These include (1) the requirement for base-pairing at most, but not all, positions within the pre-mir-451 stem; (2) surprisingly little influence of the 5'-nucleotide on Ago sorting; (3) substantial influence of Ago protein stoichiometry on mir-451 maturation; (4) strong influence of G:C content in the distal stem on 3' resection of cleaved mir-451 substrates; and (5) the influence of hairpin length on substrate utilization by Ago2 and Dicer. Unexpectedly, we find that certain hairpin lengths confer competence to mature via both Dicer-mediated and Ago2-mediated pathways, and we show, in fact, that a conventional shRNA can traverse the Dicer-independent pathway. Altogether, these data inform the design of effective Dicer-independent substrates for gene silencing and reveal novel aspects of substrate handling by Ago proteins.
Figures




References
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 - PubMed
-
- Brummelkamp TR, Bernards R, Agami R 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
