Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into lake geneva, Switzerland
- PMID: 22461783
- PMCID: PMC3310248
- DOI: 10.3389/fmicb.2012.00106
Increased levels of multiresistant bacteria and resistance genes after wastewater treatment and their dissemination into lake geneva, Switzerland
Abstract
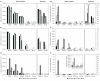
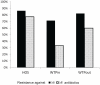
At present, very little is known about the fate and persistence of multiresistant bacteria (MRB) and their resistance genes in natural aquatic environments. Treated, but partly also untreated sewage of the city of Lausanne, Switzerland is discharged into Vidy Bay (Lake Geneva) resulting in high levels of contamination in this part of the lake. In the present work we have studied the prevalence of MRB and resistance genes in the wastewater stream of Lausanne. Samples from hospital and municipal raw sewage, treated effluent from Lausanne's wastewater treatment plant (WTP) as well as lake water and sediment samples obtained close to the WTP outlet pipe and a remote site close to a drinking water pump were evaluated for the prevalence of MRB. Selected isolates were identified (16S rRNA gene fragment sequencing) and characterized with regards to further resistances, resistance genes, and plasmids. Mostly, studies investigating this issue have relied on cultivation-based approaches. However, the limitations of these tools are well known, in particular for environmental microbial communities, and cultivation-independent molecular tools should be applied in parallel in order to take non-culturable organisms into account. Here we directly quantified the sulfonamide resistance genes sul1 and sul2 from environmental DNA extracts using TaqMan real-time quantitative PCR. Hospital sewage contained the highest load of MRB and antibiotic resistance genes (ARGs). Wastewater treatment reduced the total bacterial load up to 78% but evidence for selection of extremely multiresistant strains and accumulation of resistance genes was observed. Our data clearly indicated pollution of sediments with ARGs in the vicinity of the WTP outlet. The potential of lakes as reservoirs of MRB and potential risks are discussed.
Keywords: antibiotics; aquatic; environment; pollution; qPCR; sediment; sewage.
Figures

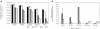


References
-
- Alder A. C., McArdell C. S., Golet E. M., Ibric S., Molnar E., Nipales Norriel S., Giger W. (2001). “Occurrence and fate of fluoroquinolone, macrolide, and sulfonamide antibiotics during wastewater treatment and in ambient waters in Switzerland,” in Pharmaceuticals and Care Products in the Environment: Scientific and Regulatory Issues, ACS Symposium Series, Vol. 791, Daughton C. G., Jones-Lepp T. L. (Washington, DC: American Chemical Society; ), 56–69
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

