Categorization of 77 dystrophin exons into 5 groups by a decision tree using indexes of splicing regulatory factors as decision markers
- PMID: 22462762
- PMCID: PMC3350383
- DOI: 10.1186/1471-2156-13-23
Categorization of 77 dystrophin exons into 5 groups by a decision tree using indexes of splicing regulatory factors as decision markers
Abstract
Background: Duchenne muscular dystrophy, a fatal muscle-wasting disease, is characterized by dystrophin deficiency caused by mutations in the dystrophin gene. Skipping of a target dystrophin exon during splicing with antisense oligonucleotides is attracting much attention as the most plausible way to express dystrophin in DMD. Antisense oligonucleotides have been designed against splicing regulatory sequences such as splicing enhancer sequences of target exons. Recently, we reported that a chemical kinase inhibitor specifically enhances the skipping of mutated dystrophin exon 31, indicating the existence of exon-specific splicing regulatory systems. However, the basis for such individual regulatory systems is largely unknown. Here, we categorized the dystrophin exons in terms of their splicing regulatory factors.
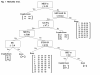
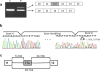
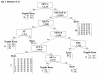
Results: Using a computer-based machine learning system, we first constructed a decision tree separating 77 authentic from 14 known cryptic exons using 25 indexes of splicing regulatory factors as decision markers. We evaluated the classification accuracy of a novel cryptic exon (exon 11a) identified in this study. However, the tree mislabeled exon 11a as a true exon. Therefore, we re-constructed the decision tree to separate all 15 cryptic exons. The revised decision tree categorized the 77 authentic exons into five groups. Furthermore, all nine disease-associated novel exons were successfully categorized as exons, validating the decision tree. One group, consisting of 30 exons, was characterized by a high density of exonic splicing enhancer sequences. This suggests that AOs targeting splicing enhancer sequences would efficiently induce skipping of exons belonging to this group.
Conclusions: The decision tree categorized the 77 authentic exons into five groups. Our classification may help to establish the strategy for exon skipping therapy for Duchenne muscular dystrophy.
Figures
References
-
- Nishio H, Takeshima Y, Narita N, Yanagawa H, Suzuki Y, Ishikawa Y, Minami R, Nakamura H, Matsuo M. Identification of a novel first exon in the human dystrophin gene and of a new promoter located more than 500 kb upstream of the nearest known promoter. J Clin Invest. 1994;94:1037–1042. doi: 10.1172/JCI117417. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

