Small non-coding RNAs mount a silent revolution in gene expression
- PMID: 22464106
- PMCID: PMC3372702
- DOI: 10.1016/j.ceb.2012.03.006
Small non-coding RNAs mount a silent revolution in gene expression
Abstract
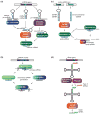
During the past decade, it has become evident that small non-coding RNAs (ncRNAs) participate in widespread and essential regulatory mechanisms in most eukaryotic cells. Novel classes of small RNAs, their biogenesis pathways and cellular effects are continuously being described, and new properties of already established ncRNAs are still being discovered. As the list of small RNA molecules and their roles becomes more and more extensive, one can get lost in the midst of new information. In this review, we attempt to bring order to the small ncRNA transcriptome by covering some of the major milestones of recent years. We go through many of the new properties that have been attributed to already familiar RNA molecules, and introduce some of the more recent novel classes of tiny ncRNAs.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

References
-
- Vazquez F, Legrand S, Windels D. The biosynthetic pathways and biological scopes of plant small RNAs. Trends Plant Sci. 2010;15:337–345. - PubMed
-
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. - PubMed
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. - PubMed
-
- Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. These three papers reveal a novel miRNA processing pathway by showing that the maturation of pre-miR-451 does not require Dicer processing but is dependent on the catalytic activity of Ago2. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

