Chlamydia trachomatis Tarp cooperates with the Arp2/3 complex to increase the rate of actin polymerization
- PMID: 22465117
- PMCID: PMC3334425
- DOI: 10.1016/j.bbrc.2012.03.080
Chlamydia trachomatis Tarp cooperates with the Arp2/3 complex to increase the rate of actin polymerization
Abstract
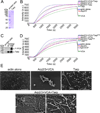
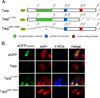
Actin polymerization is required for Chlamydia trachomatis entry into nonphagocytic host cells. Host and chlamydial actin nucleators are essential for internalization of chlamydiae by eukaryotic cells. The host cell Arp2/3 complex and the chlamydial translocated actin recruiting phosphoprotein (Tarp) are both required for entry. Tarp and the Arp2/3 complex exhibit unique actin polymerization kinetics individually, but the molecular details of how these two actin nucleators cooperate to promote bacterial entry is not understood. In this study we provide biochemical evidence that the two actin nucleators act synergistically by co-opting the unique attributes of each to enhance the dynamics of actin filament formation. This process is independent of Tarp phosphorylation. We further demonstrate that Tarp colocalization with actin filaments is independent of the Tarp phosphorylation domain. The results are consistent with a model in which chlamydial and host cell actin nucleators cooperate to increase the rate of actin filament formation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
References
-
- CDC Grand Rounds: Chlamydia prevention: challenges and strategies for reducing disease burden and sequelae. MMWR Morb Mortal Wkly Rep. 60:370–373. - PubMed
-
- Schachter J. Infection and disease epidemiology. In: Stephens RS, editor. Chlamydia; Intracellular biology, pathogenesis, and immunity. Washington, D.C.: ASM Press; 1999. pp. 139–169.
-
- Mariotti SP, Pascolini D, Rose-Nussbaumer J. Trachoma: global magnitude of a preventable cause of blindness. Br J Ophthalmol. 2009;93:563–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

