Homologous recombination and its regulation
- PMID: 22467216
- PMCID: PMC3401455
- DOI: 10.1093/nar/gks270
Homologous recombination and its regulation
Abstract
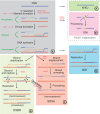
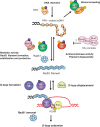
Homologous recombination (HR) is critical both for repairing DNA lesions in mitosis and for chromosomal pairing and exchange during meiosis. However, some forms of HR can also lead to undesirable DNA rearrangements. Multiple regulatory mechanisms have evolved to ensure that HR takes place at the right time, place and manner. Several of these impinge on the control of Rad51 nucleofilaments that play a central role in HR. Some factors promote the formation of these structures while others lead to their disassembly or the use of alternative repair pathways. In this article, we review these mechanisms in both mitotic and meiotic environments and in different eukaryotic taxa, with an emphasis on yeast and mammal systems. Since mutations in several proteins that regulate Rad51 nucleofilaments are associated with cancer and cancer-prone syndromes, we discuss how understanding their functions can lead to the development of better tools for cancer diagnosis and therapy.
Figures
References
-
- Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003;278:42729–42732. - PubMed
-
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. - PubMed
-
- Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

