Cellular quantitative structure-activity relationship (Cell-QSAR): conceptual dissection of receptor binding and intracellular disposition in antifilarial activities of Selwood antimycins
- PMID: 22468611
- PMCID: PMC3338160
- DOI: 10.1021/jm201371y
Cellular quantitative structure-activity relationship (Cell-QSAR): conceptual dissection of receptor binding and intracellular disposition in antifilarial activities of Selwood antimycins
Abstract
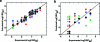
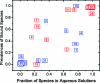
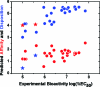
We present the cellular quantitative structure-activity relationship (cell-QSAR) concept that adapts ligand-based and receptor-based 3D-QSAR methods for use with cell-level activities. The unknown intracellular drug disposition is accounted for by the disposition function (DF), a model-based, nonlinear function of a drug's lipophilicity, acidity, and other properties. We conceptually combined the DF with our multispecies, multimode version of the frequently used ligand-based comparative molecular field analysis (CoMFA) method, forming a single correlation function for fitting the cell-level activities. The resulting cell-QSAR model was applied to the Selwood data on filaricidal activities of antimycin analogues. Their molecules are flexible, ionize under physiologic conditions, form different intramolecular H-bonds for neutral and ionized species, and cross several membranes to reach unknown receptors. The calibrated cell-QSAR model is significantly more predictive than other models lacking the disposition part and provides valuable structure optimization clues by factorizing the cell-level activity of each compound into the contributions of the receptor binding and disposition.
Figures





Similar articles
-
Rigorous treatment of multispecies multimode ligand-receptor interactions in 3D-QSAR: CoMFA analysis of thyroxine analogs binding to transthyretin.J Chem Inf Model. 2011 May 23;51(5):1132-50. doi: 10.1021/ci200055s. Epub 2011 Apr 8. J Chem Inf Model. 2011. PMID: 21476521 Free PMC article.
-
Exploration of a binding mode of indole amide analogues as potent histone deacetylase inhibitors and 3D-QSAR analyses.Bioorg Med Chem. 2005 Sep 15;13(18):5424-34. doi: 10.1016/j.bmc.2005.05.016. Bioorg Med Chem. 2005. PMID: 15963726
-
Computational Analysis of CRTh2 receptor antagonist: A Ligand-based CoMFA and CoMSIA approach.Comput Biol Chem. 2015 Jun;56:109-21. doi: 10.1016/j.compbiolchem.2015.04.007. Epub 2015 Apr 20. Comput Biol Chem. 2015. PMID: 25935115
-
Development of quantitative structure-activity relationships and its application in rational drug design.Curr Pharm Des. 2006;12(35):4601-11. doi: 10.2174/138161206779010431. Curr Pharm Des. 2006. PMID: 17168765 Review.
-
3D-QSAR in drug design--a review.Curr Top Med Chem. 2010;10(1):95-115. doi: 10.2174/156802610790232260. Curr Top Med Chem. 2010. PMID: 19929826 Review.
Cited by
-
Web-Based Quantitative Structure-Activity Relationship Resources Facilitate Effective Drug Discovery.Top Curr Chem (Cham). 2021 Sep 23;379(6):37. doi: 10.1007/s41061-021-00349-3. Top Curr Chem (Cham). 2021. PMID: 34554348 Review.
-
Structure-based prediction of drug distribution across the headgroup and core strata of a phospholipid bilayer using surrogate phases.Mol Pharm. 2014 Oct 6;11(10):3577-95. doi: 10.1021/mp5003366. Epub 2014 Sep 18. Mol Pharm. 2014. PMID: 25179490 Free PMC article.
-
Biomacromolecular quantitative structure-activity relationship (BioQSAR): a proof-of-concept study on the modeling, prediction and interpretation of protein-protein binding affinity.J Comput Aided Mol Des. 2013 Jan;27(1):67-78. doi: 10.1007/s10822-012-9625-3. Epub 2013 Jan 10. J Comput Aided Mol Des. 2013. PMID: 23306464
-
Does transbilayer diffusion have a role in membrane transport of drugs?Drug Discov Today. 2012 Oct;17(19-20):1079-87. doi: 10.1016/j.drudis.2012.06.003. Epub 2012 Jun 15. Drug Discov Today. 2012. PMID: 22705388 Free PMC article. Review.
-
Rigorous incorporation of tautomers, ionization species, and different binding modes into ligand-based and receptor-based 3D-QSAR methods.Curr Pharm Des. 2013;19(23):4316-22. doi: 10.2174/1381612811319230013. Curr Pharm Des. 2013. PMID: 23170882 Free PMC article. Review.
References
-
- Gilson M. K.; Zhou H. X. Calculation of Protein-Ligand Binding Affinities. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 21–42. - PubMed
-
- Kollman P. Free Energy Calculations: Applications to Chemical and Biochemical Phenomena. Chem. Rev. 1993, 93, 2395–2417.
-
- Aqvist J.; Medina C.; Samuelsson J. E. A New Method for Predicting Binding Affinity in Computer-Aided Drug Design. Protein Eng. 1994, 7, 385–391. - PubMed
-
- Hansson T.; Aqvist J. Estimation of Binding Free Energies for HIV Proteinase Inhibitors by Molecular Dynamics Simulations. Protein Eng. 1995, 8, 1137–1144. - PubMed
-
- Aqvist J. Calculation of Absolute Binding Free Energies for Charged Ligands and Effects of Long-Range Electrostatic Interactions. J. Comput. Chem. 1996, 17, 1587–1597.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

