Henipavirus mediated membrane fusion, virus entry and targeted therapeutics
- PMID: 22470837
- PMCID: PMC3315217
- DOI: 10.3390/v4020280
Henipavirus mediated membrane fusion, virus entry and targeted therapeutics
Abstract
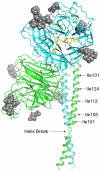
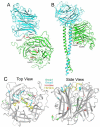
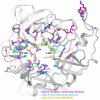
The Paramyxoviridae genus Henipavirus is presently represented by the type species Hendra and Nipah viruses which are both recently emerged zoonotic viral pathogens responsible for repeated outbreaks associated with high morbidity and mortality in Australia, Southeast Asia, India and Bangladesh. These enveloped viruses bind and enter host target cells through the coordinated activities of their attachment (G) and class I fusion (F) envelope glycoproteins. The henipavirus G glycoprotein interacts with host cellular B class ephrins, triggering conformational alterations in G that lead to the activation of the F glycoprotein, which facilitates the membrane fusion process. Using the recently published structures of HeV-G and NiV-G and other paramyxovirus glycoproteins, we review the features of the henipavirus envelope glycoproteins that appear essential for mediating the viral fusion process, including receptor binding, G-F interaction, F activation, with an emphasis on G and the mutations that disrupt viral infectivity. Finally, recent candidate therapeutics for henipavirus-mediated disease are summarized in light of their ability to inhibit HeV and NiV entry by targeting their G and F glycoproteins.
Keywords: antibody; attachment glycoprotein; entry; ephrin-B2; ephrin-B3; fusion glycoprotein; immunotherapy; inhibitor; membrane fusion; paramyxovirus; receptor.
Figures
References
-
- Selvey L.A., Wells R.M., McCormack J.G., Ansford A.J., Murray K., Rogers R.J., Lavercombe P.S., Selleck P., Sheridan J.W. Infection of humans and horses by a newly described morbillivirus [see comments]. Med. J. Aust. 1995;162:642–645. - PubMed
-
- Chua K.B., Goh K.J., Wong K.T., Kamarulzaman A., Tan P.S., Ksiazek T.G., Zaki S.R., Paul G., Lam S.K., Tan C.T. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia [see comments]. Lancet. 1999;354:1257–1259. - PubMed
-
- Chua K.B., Bellini W.J., Rota P.A., Harcourt B.H., Tamin A., Lam S.K., Ksiazek T.G., Rollin P.E., Zaki S.R., Shieh W., et al. Nipah virus: A recently emergent deadly paramyxovirus. Science. 2000;288:1432–1435. - PubMed
-
- Chong H.T., Kunjapan S.R., Thayaparan T., Tong J., Petharunam V., Jusoh M.R., Tan C.T. Nipah encephalitis outbreak in Malaysia, clinical features in patients from Seremban. Can. J. Neurol. Sci. 2002;29:83–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

