Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve
- PMID: 22471618
- PMCID: PMC3372441
- DOI: 10.1186/1471-2148-12-46
Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve
Abstract
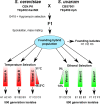
Background: Interspecific hybridization occurs in every eukaryotic kingdom. While hybrid progeny are frequently at a selective disadvantage, in some instances their increased genome size and complexity may result in greater stress resistance than their ancestors, which can be adaptively advantageous at the edges of their ancestors' ranges. While this phenomenon has been repeatedly documented in the field, the response of hybrid populations to long-term selection has not often been explored in the lab. To fill this knowledge gap we crossed the two most distantly related members of the Saccharomyces sensu stricto group, S. cerevisiae and S. uvarum, and established a mixed population of homoploid and aneuploid hybrids to study how different types of selection impact hybrid genome structure.
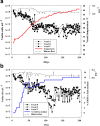
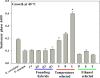
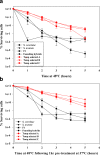
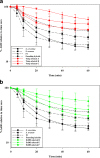
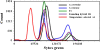
Results: As temperature was raised incrementally from 31°C to 46.5°C over 500 generations of continuous culture, selection favored loss of the S. uvarum genome, although the kinetics of genome loss differed among independent replicates. Temperature-selected isolates exhibited greater inherent and induced thermal tolerance than parental species and founding hybrids, and also exhibited ethanol resistance. In contrast, as exogenous ethanol was increased from 0% to 14% over 500 generations of continuous culture, selection favored euploid S. cerevisiae x S. uvarum hybrids. Ethanol-selected isolates were more ethanol tolerant than S. uvarum and one of the founding hybrids, but did not exhibit resistance to temperature stress. Relative to parental and founding hybrids, temperature-selected strains showed heritable differences in cell wall structure in the forms of increased resistance to zymolyase digestion and Micafungin, which targets cell wall biosynthesis.
Conclusions: This is the first study to show experimentally that the genomic fate of newly-formed interspecific hybrids depends on the type of selection they encounter during the course of evolution, underscoring the importance of the ecological theatre in determining the outcome of the evolutionary play.
Figures


Similar articles
-
Temperature preference can bias parental genome retention during hybrid evolution.PLoS Genet. 2019 Sep 16;15(9):e1008383. doi: 10.1371/journal.pgen.1008383. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31525194 Free PMC article.
-
Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae.FEMS Yeast Res. 2005 Dec;5(12):1141-50. doi: 10.1016/j.femsyr.2005.04.008. Epub 2005 Jun 15. FEMS Yeast Res. 2005. PMID: 15982931
-
Genotypic and phenotypic evolution of yeast interspecies hybrids during high-sugar fermentation.Appl Microbiol Biotechnol. 2016 Jul;100(14):6331-6343. doi: 10.1007/s00253-016-7481-0. Epub 2016 Apr 13. Appl Microbiol Biotechnol. 2016. PMID: 27075738
-
Interspecies hybridization and recombination in Saccharomyces wine yeasts.FEMS Yeast Res. 2008 Nov;8(7):996-1007. doi: 10.1111/j.1567-1364.2008.00369.x. Epub 2008 Mar 18. FEMS Yeast Res. 2008. PMID: 18355270 Review.
-
Chapter 6: The genomes of lager yeasts.Adv Appl Microbiol. 2009;69:159-82. doi: 10.1016/S0065-2164(09)69006-7. Adv Appl Microbiol. 2009. PMID: 19729094 Review.
Cited by
-
Aneuploidy in yeast: Segregation error or adaptation mechanism?Yeast. 2019 Sep;36(9):525-539. doi: 10.1002/yea.3427. Epub 2019 Aug 1. Yeast. 2019. PMID: 31199875 Free PMC article.
-
Yeast two- and three-species hybrids and high-sugar fermentation.Microb Biotechnol. 2019 Nov;12(6):1101-1108. doi: 10.1111/1751-7915.13390. Epub 2019 Mar 5. Microb Biotechnol. 2019. PMID: 30838806 Free PMC article. Review.
-
Experimental evolution of protein-protein interaction networks.Biochem J. 2013 Aug 1;453(3):311-9. doi: 10.1042/BJ20130205. Biochem J. 2013. PMID: 23849056 Free PMC article. Review.
-
Interspecies Hybridisation and Genome Chimerisation in Saccharomyces: Combining of Gene Pools of Species and Its Biotechnological Perspectives.Front Microbiol. 2018 Dec 11;9:3071. doi: 10.3389/fmicb.2018.03071. eCollection 2018. Front Microbiol. 2018. PMID: 30619156 Free PMC article. Review.
-
A Unique Saccharomyces cerevisiae × Saccharomyces uvarum Hybrid Isolated From Norwegian Farmhouse Beer: Characterization and Reconstruction.Front Microbiol. 2018 Sep 24;9:2253. doi: 10.3389/fmicb.2018.02253. eCollection 2018. Front Microbiol. 2018. PMID: 30319573 Free PMC article.
References
-
- Sexton JP, McIntyre PJ, Angert AL, Rice KJ. Evolution and ecology of species range limits. Ann Rev Ecol Evol Systemat. 2009;40:415–436. doi: 10.1146/annurev.ecolsys.110308.120317. - DOI
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

