Putative resistance gene markers associated with quantitative trait loci for fire blight resistance in Malus 'Robusta 5' accessions
- PMID: 22471693
- PMCID: PMC3443455
- DOI: 10.1186/1471-2156-13-25
Putative resistance gene markers associated with quantitative trait loci for fire blight resistance in Malus 'Robusta 5' accessions
Abstract
Background: Breeding of fire blight resistant scions and rootstocks is a goal of several international apple breeding programs, as options are limited for management of this destructive disease caused by the bacterial pathogen Erwinia amylovora. A broad, large-effect quantitative trait locus (QTL) for fire blight resistance has been reported on linkage group 3 of Malus 'Robusta 5'. In this study we identified markers derived from putative fire blight resistance genes associated with the QTL by integrating further genetic mapping studies with bioinformatics analysis of transcript profiling data and genome sequence databases.
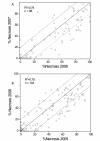
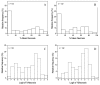
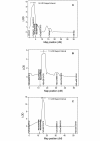
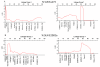
Results: When several defined E.amylovora strains were used to inoculate three progenies from international breeding programs, all with 'Robusta 5' as a common parent, two distinct QTLs were detected on linkage group 3, where only one had previously been mapped. In the New Zealand 'Malling 9' X 'Robusta 5' population inoculated with E. amylovora ICMP11176, the proximal QTL co-located with SNP markers derived from a leucine-rich repeat, receptor-like protein (MxdRLP1) and a closely linked class 3 peroxidase gene. While the QTL detected in the German 'Idared' X 'Robusta 5' population inoculated with E. amylovora strains Ea222_JKI or ICMP11176 was approximately 6 cM distal to this, directly below a SNP marker derived from a heat shock 90 family protein gene (HSP90). In the US 'Otawa3' X 'Robusta5' population inoculated with E. amylovora strains Ea273 or E2002a, the position of the LOD score peak on linkage group 3 was dependent upon the pathogen strains used for inoculation. One of the five MxdRLP1 alleles identified in fire blight resistant and susceptible cultivars was genetically associated with resistance and used to develop a high resolution melting PCR marker. A resistance QTL detected on linkage group 7 of the US population co-located with another HSP90 gene-family member and a WRKY transcription factor previously associated with fire blight resistance. However, this QTL was not observed in the New Zealand or German populations.
Conclusions: The results suggest that the upper region of 'Robusta 5' linkage group 3 contains multiple genes contributing to fire blight resistance and that their contributions to resistance can vary depending upon pathogen virulence and other factors. Mapping markers derived from putative fire blight resistance genes has proved a useful aid in defining these QTLs and developing markers for marker-assisted breeding of fire blight resistance.
Figures


Similar articles
-
Mapping of fire blight resistance in Malus ×robusta 5 flowers following artificial inoculation.BMC Plant Biol. 2019 Dec 2;19(1):532. doi: 10.1186/s12870-019-2154-7. BMC Plant Biol. 2019. PMID: 31791233 Free PMC article.
-
Malus Hosts-Erwinia amylovora Interactions: Strain Pathogenicity and Resistance Mechanisms.Front Plant Sci. 2019 Apr 26;10:551. doi: 10.3389/fpls.2019.00551. eCollection 2019. Front Plant Sci. 2019. PMID: 31105734 Free PMC article. Review.
-
Mapping of quantitative trait loci for fire blight resistance in the apple cultivars 'Florina' and 'Nova Easygro'.Genome. 2010 Sep;53(9):710-22. doi: 10.1139/g10-047. Genome. 2010. PMID: 20924420
-
Identification of genetic loci associated with fire blight resistance in Malus through combined use of QTL and association mapping.Physiol Plant. 2013 Jul;148(3):344-53. doi: 10.1111/ppl.12068. Epub 2013 May 24. Physiol Plant. 2013. PMID: 23627651
-
Fire blight: applied genomic insights of the pathogen and host.Annu Rev Phytopathol. 2012;50:475-94. doi: 10.1146/annurev-phyto-081211-172931. Epub 2012 Jun 11. Annu Rev Phytopathol. 2012. PMID: 22702352 Review.
Cited by
-
Construction of a dense genetic map of the Malus fusca fire blight resistant accession MAL0045 using tunable genotyping-by-sequencing SNPs and microsatellites.Sci Rep. 2020 Oct 1;10(1):16358. doi: 10.1038/s41598-020-73393-6. Sci Rep. 2020. PMID: 33005026 Free PMC article.
-
Mapping of fire blight resistance in Malus ×robusta 5 flowers following artificial inoculation.BMC Plant Biol. 2019 Dec 2;19(1):532. doi: 10.1186/s12870-019-2154-7. BMC Plant Biol. 2019. PMID: 31791233 Free PMC article.
-
Malus Hosts-Erwinia amylovora Interactions: Strain Pathogenicity and Resistance Mechanisms.Front Plant Sci. 2019 Apr 26;10:551. doi: 10.3389/fpls.2019.00551. eCollection 2019. Front Plant Sci. 2019. PMID: 31105734 Free PMC article. Review.
-
The Arabidopsis pattern recognition receptor EFR enhances fire blight resistance in apple.Hortic Res. 2021 Sep 1;8(1):204. doi: 10.1038/s41438-021-00639-3. Hortic Res. 2021. PMID: 34465763 Free PMC article.
-
Fire blight disease reactome: RNA-seq transcriptional profile of apple host plant defense responses to Erwinia amylovora pathogen infection.Sci Rep. 2016 Feb 17;6:21600. doi: 10.1038/srep21600. Sci Rep. 2016. PMID: 26883568 Free PMC article.
References
-
- Griffith CS, Sutton TB, Peterson PD, editor. Fire blight, The Foundation of Phytobacteriology. APS Press, St Paul, Minn; 2003.
-
- Peil A, Bus VGM, Geider K, Richter K, Flachowsky H, Hanke M-V. Improvement of fire blight resistance in apple and pear. Int J Plant Breed. 2009;3(1):1–27. doi: 10.3923/ijpbg.2009.1.10. - DOI
-
- Roberts RG, Sawyer AJ. An updated pest risk assessment for spread ofErwinia amylovoraand fire blight via commercial apple fruit. Crop Prot. 2008;27:362–368. doi: 10.1016/j.cropro.2007.06.007. - DOI
-
- USDA-National Institute of Food and Agriculture, Plant Breeding, Genetics, Genomics, Fire Blight Resistant Apples. http://www.csrees.usda.gov/nea/plants/in_focus/pbgg_if_fire_blight_resis...
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

