Deep sequencing the circadian and diurnal transcriptome of Drosophila brain
- PMID: 22472103
- PMCID: PMC3396368
- DOI: 10.1101/gr.128876.111
Deep sequencing the circadian and diurnal transcriptome of Drosophila brain
Abstract
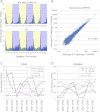
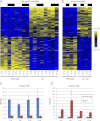
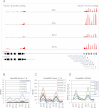
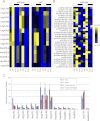
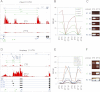
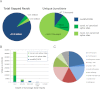
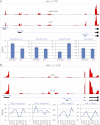
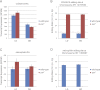
Eukaryotic circadian clocks include transcriptional/translational feedback loops that drive 24-h rhythms of transcription. These transcriptional rhythms underlie oscillations of protein abundance, thereby mediating circadian rhythms of behavior, physiology, and metabolism. Numerous studies over the last decade have used microarrays to profile circadian transcriptional rhythms in various organisms and tissues. Here we use RNA sequencing (RNA-seq) to profile the circadian transcriptome of Drosophila melanogaster brain from wild-type and period-null clock-defective animals. We identify several hundred transcripts whose abundance oscillates with 24-h periods in either constant darkness or 12 h light/dark diurnal cycles, including several noncoding RNAs (ncRNAs) that were not identified in previous microarray studies. Of particular interest are U snoRNA host genes (Uhgs), a family of diurnal cycling noncoding RNAs that encode the precursors of more than 50 box-C/D small nucleolar RNAs, key regulators of ribosomal biogenesis. Transcriptional profiling at the level of individual exons reveals alternative splice isoforms for many genes whose relative abundances are regulated by either period or circadian time, although the effect of circadian time is muted in comparison to that of period. Interestingly, period loss of function significantly alters the frequency of RNA editing at several editing sites, suggesting an unexpected link between a key circadian gene and RNA editing. We also identify tens of thousands of novel splicing events beyond those previously annotated by the modENCODE Consortium, including several that affect key circadian genes. These studies demonstrate extensive circadian control of ncRNA expression, reveal the extent of clock control of alternative splicing and RNA editing, and provide a novel, genome-wide map of splicing in Drosophila brain.
Figures

References
-
- Bass BL. 2001. RNA editing. Oxford University Press, New York.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
