Functional repertoire, molecular pathways and diseases associated with 3D domain swapping in the human proteome
- PMID: 22472218
- PMCID: PMC3508620
- DOI: 10.1186/2043-9113-2-8
Functional repertoire, molecular pathways and diseases associated with 3D domain swapping in the human proteome
Abstract
Background: 3D domain swapping is a novel structural phenomenon observed in diverse set of protein structures in oligomeric conformations. A distinct structural feature, where structural segments in a protein dimer or higher oligomer were shared between two or more chains of a protein structure, characterizes 3D domain swapping. 3D domain swapping was observed as a key mediator of numerous functional mechanisms and play pathogenic role in various diseases including conformational diseases like amyloidosis, Alzheimer's disease, Parkinson's disease and prion diseases. We report the first study with a focus on identifying functional classes, pathways and diseases mediated by 3D domain swapping in the human proteome.
Methods: We used a panel of four enrichment tools with two different ontologies and two annotations database to derive biological and clinical relevant information associated with 3D domain swapping. Protein domain enrichment analysis followed by Gene Ontology (GO) term enrichment analysis revealed the functional repertoire of proteins involved in swapping. Pathway analysis using KEGG annotations revealed diverse pathway associations of human proteins involved in 3D domain swapping. Disease Ontology was used to find statistically significant associations with proteins in swapped conformation and various disease categories (P-value < 0.05).
Results: We report meta-analysis results of a literature-curated dataset of human gene products involved in 3D domain swapping and discuss new insights about the functional repertoire, pathway associations and disease implications of proteins involved in 3D domain swapping.
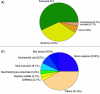
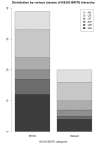
Conclusions: Our integrated bioinformatics pipeline comprising of four different enrichment tools, two ontologies and two annotations revealed new insights into the functional and disease correlations with 3D domain swapping. GO term enrichment were used to infer terms associated with three different GO categories. Protein domain enrichment was used to identify conserved domains enriched in swapped proteins. Pathway enrichment analysis using KEGG annotations revealed that proteins with swapped conformations are present in all six classes of KEGG BRITE hierarchy and significantly enriched KEGG pathways were observed in five classes. Five major classes of disease were found to be associated with 3D domain swapping using functional disease ontology based enrichment analysis. Five classes of human diseases: cancer, diseases of the respiratory or pulmonary system, degenerative diseases of the central nervous system, vascular disease and encephalitis were found to be significant. In conclusion, our study shows that bioinformatics based analytical approaches using curated data can enhance the understanding of functional and disease implications of 3D domain swapping.
Figures

Similar articles
-
3DSwap: curated knowledgebase of proteins involved in 3D domain swapping.Database (Oxford). 2011 Sep 29;2011:bar042. doi: 10.1093/database/bar042. Print 2011. Database (Oxford). 2011. PMID: 21959866 Free PMC article.
-
3D domain swapping: as domains continue to swap.Protein Sci. 2002 Jun;11(6):1285-99. doi: 10.1110/ps.0201402. Protein Sci. 2002. PMID: 12021428 Free PMC article. Review.
-
Genome-Wide Prediction and Analysis of 3D-Domain Swapped Proteins in the Human Genome from Sequence Information.PLoS One. 2016 Jul 28;11(7):e0159627. doi: 10.1371/journal.pone.0159627. eCollection 2016. PLoS One. 2016. PMID: 27467780 Free PMC article.
-
Insights into Protein Sequence and Structure-Derived Features Mediating 3D Domain Swapping Mechanism using Support Vector Machine Based Approach.Bioinform Biol Insights. 2010 Jun 17;4:33-42. doi: 10.4137/bbi.s4464. Bioinform Biol Insights. 2010. PMID: 20634983 Free PMC article.
-
3D domain swapping: a mechanism for oligomer assembly.Protein Sci. 1995 Dec;4(12):2455-68. doi: 10.1002/pro.5560041202. Protein Sci. 1995. PMID: 8580836 Free PMC article. Review.
Cited by
-
Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning.Brief Bioinform. 2018 Jul 20;19(4):656-678. doi: 10.1093/bib/bbw136. Brief Bioinform. 2018. PMID: 28200013 Free PMC article.
-
Linking in domain-swapped protein dimers.Sci Rep. 2016 Sep 23;6:33872. doi: 10.1038/srep33872. Sci Rep. 2016. PMID: 27659606 Free PMC article.
-
Editorial: Improving Neuropharmacology using Big Data, Machine Learning and Computational Algorithms.Curr Neuropharmacol. 2017 Nov 14;15(8):1058-1061. doi: 10.2174/1570159X1508171114113425. Curr Neuropharmacol. 2017. PMID: 29199918 Free PMC article. No abstract available.
-
Genome mining the black-yeast Aureobasidium pullulans NRRL 62031 for biotechnological traits.BMC Genomics. 2025 Mar 13;26(1):244. doi: 10.1186/s12864-025-11395-2. BMC Genomics. 2025. PMID: 40082747 Free PMC article.
-
An integrative pipeline for multi-modal discovery of disease relationships.Pac Symp Biocomput. 2015;20:407-18. Pac Symp Biocomput. 2015. PMID: 25592600 Free PMC article.
References
-
- Reddy Chilamakuri CS, Sekhar SK, Bernard Offmann, Sowdhamini Ramanathan. PURE: a web server for querying the relationship between pre-existing domains and unassigned regions in proteins. Nature Protocol Exchange. 2007. doi:10.1038/nprot.2007.486.
-
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. - PubMed
LinkOut - more resources
Full Text Sources

