The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer
- PMID: 22472886
- PMCID: PMC3341860
- DOI: 10.1038/bjc.2012.109
The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer
Erratum in
-
The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer.Br J Cancer. 2017 Aug 22;117(5):e2. doi: 10.1038/bjc.2017.203. Epub 2017 Jul 13. Br J Cancer. 2017. PMID: 28704839 Free PMC article.
Abstract
Background: Although the role of human papilloma virus (HPV) in cervical squamous cell carcinoma (CSCC) is well established, the role in head and neck SCC (HNSCC) is less clear. MicroRNAs (miRNAs) have a role in the cancer development, and HPV status may affect the miRNA expression pattern in HNSCC. To explore the influence of HPV in HNSCC, we made a comparative miRNA profile of HPV-positive (HPV+) and HPV-negative (HPV-) HNSCC against CSCC.
Methods: Fresh frozen and laser microdissected-paraffin-embedded samples obtained from patients with HPV+/HPV- HNSCC, CSCC and controls were used for microarray analysis. Differentially expressed miRNAs in the HPV+ and HPV- HNSCC samples were compared with the differentially expressed miRNAs in the CSCC samples.
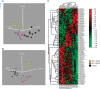
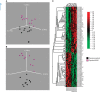
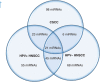
Results: Human papilloma virus positive (+) HNSCC had a distinct miRNA profile compared with HPV- HNSCC. Significantly more similarity was seen between HPV+ HNSCC and CSCC than HPV- and CSCC. A set of HPV core miRNAs were identified. Of these especially the miR-15a/miR-16/miR195/miR-497 family, miR-143/miR-145 and the miR-106-363 cluster appear to be important within the known HPV pathogenesis.
Conclusion: This study adds new knowledge to the known pathogenic pathways of HPV and substantiates the oncogenic role of HPV in subsets of HNSCCs.
Figures

Similar articles
-
MicroRNAs as new biomarkers for human papilloma virus related head and neck cancers.Cancer Biomark. 2015;15(3):213-8. doi: 10.3233/CBM-150464. Cancer Biomark. 2015. PMID: 25769448 Review.
-
Genome-wide miRNA profiling reinforces the importance of miR-9 in human papillomavirus associated oral and oropharyngeal head and neck cancer.Sci Rep. 2019 Feb 19;9(1):2306. doi: 10.1038/s41598-019-38797-z. Sci Rep. 2019. PMID: 30783190 Free PMC article.
-
Expression of microRNAs in squamous cell carcinoma of human head and neck and the esophagus: miR-205 and miR-21 are specific markers for HNSCC and ESCC.Oncol Rep. 2010 Jun;23(6):1625-33. doi: 10.3892/or_00000804. Oncol Rep. 2010. PMID: 20428818
-
DNA methylation regulated microRNAs in HPV-16-induced head and neck squamous cell carcinoma (HNSCC).Mol Cell Biochem. 2018 Nov;448(1-2):321-333. doi: 10.1007/s11010-018-3336-6. Epub 2018 Feb 17. Mol Cell Biochem. 2018. PMID: 29455435
-
An Update on Cellular MicroRNA Expression in Human Papillomavirus-Associated Head and Neck Squamous Cell Carcinoma.Oncology. 2018;95(4):193-201. doi: 10.1159/000489786. Epub 2018 Jun 19. Oncology. 2018. PMID: 29920485 Review.
Cited by
-
Nip the HPV encoded evil in the cancer bud: HPV reshapes TRAILs and signaling landscapes.Cancer Cell Int. 2013 Jun 17;13(1):61. doi: 10.1186/1475-2867-13-61. Cancer Cell Int. 2013. PMID: 23773282 Free PMC article.
-
Genomic landscape of human papillomavirus-associated cancers.Clin Cancer Res. 2015 May 1;21(9):2009-19. doi: 10.1158/1078-0432.CCR-14-1101. Epub 2015 Mar 16. Clin Cancer Res. 2015. PMID: 25779941 Free PMC article. Review.
-
Head and Neck Cancers Are Not Alike When Tarred with the Same Brush: An Epigenetic Perspective from the Cancerization Field to Prognosis.Cancers (Basel). 2021 Nov 11;13(22):5630. doi: 10.3390/cancers13225630. Cancers (Basel). 2021. PMID: 34830785 Free PMC article. Review.
-
Tumor microenvironment: an evil nexus promoting aggressive head and neck squamous cell carcinoma and avenue for targeted therapy.Signal Transduct Target Ther. 2021 Jan 12;6(1):12. doi: 10.1038/s41392-020-00419-w. Signal Transduct Target Ther. 2021. PMID: 33436555 Free PMC article. Review.
-
Interplay of miR-542, miR-126, miR-143 and miR-26b with PI3K-Akt is a Diagnostic Signal and Putative Regulatory Target in HPV-Positive Cervical Cancer.Biochem Genet. 2025 Jun;63(3):2760-2780. doi: 10.1007/s10528-024-10837-y. Epub 2024 Jun 7. Biochem Genet. 2025. PMID: 38849709
References
-
- Akao Y, Nakagawa Y, Naoe T (2007b) MicroRNA-143 and -145 in colon cancer. DNA Cell Biol 26(5): 311–320 - PubMed
-
- Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E (2009) miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res 69(13): 5553–5559 - PubMed
-
- Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP, Roder K, Babraj J, Wahlestedt C, Hutvagner G, Pedersen BK, Timmons JA (2010) Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med 2(2): 9. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

