Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes
- PMID: 22473038
- PMCID: PMC3427768
- DOI: 10.1038/ni.2247
Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes
Abstract
Innate γδ T cells function in the early phase of immune responses. Although innate γδ T cells have often been studied as one homogenous population, they can be functionally classified into effector subsets on the basis of the production of signature cytokines, analogous to adaptive helper T cell subsets. However, unlike the function of adaptive T cells, γδ effector T cell function correlates with genomically encoded T cell antigen receptor (TCR) chains, which suggests that clonal TCR selection is not the main determinant of the differentiation of γδ effector cells. A high-resolution transcriptome analysis of all emergent γδ thymocyte subsets segregated on the basis of use of the TCR γ-chain or δ-chain indicated the existence of three separate subtypes of γδ effector cells in the thymus. The immature γδ subsets were distinguished by unique transcription-factor modules that program effector function.
Figures

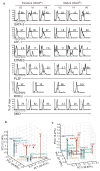
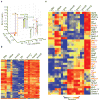
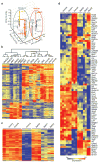
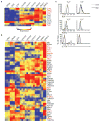

Comment in
-
Thymic signatures of tailored peripheral functions.Nat Immunol. 2012 Apr 18;13(5):431-3. doi: 10.1038/ni.2287. Nat Immunol. 2012. PMID: 22513325 No abstract available.
Similar articles
-
γδTCR-independent origin of neonatal γδ T cells prewired for IL-17 production.Curr Opin Immunol. 2019 Jun;58:60-67. doi: 10.1016/j.coi.2019.04.011. Epub 2019 May 23. Curr Opin Immunol. 2019. PMID: 31128446 Free PMC article. Review.
-
γδ T cells acquire effector fates in the thymus and differentiate into cytokine-producing effectors in a Listeria model of infection independently of CD28 costimulation.PLoS One. 2013 May 9;8(5):e63178. doi: 10.1371/journal.pone.0063178. Print 2013. PLoS One. 2013. PMID: 23671671 Free PMC article.
-
CCR6 and NK1.1 distinguish between IL-17A and IFN-gamma-producing gammadelta effector T cells.Eur J Immunol. 2009 Dec;39(12):3488-97. doi: 10.1002/eji.200939922. Eur J Immunol. 2009. PMID: 19830744
-
Lineage divergence at the first TCR-dependent checkpoint: preferential γδ and impaired αβ T cell development in nonobese diabetic mice.J Immunol. 2011 Jan 15;186(2):826-37. doi: 10.4049/jimmunol.1002630. Epub 2010 Dec 10. J Immunol. 2011. PMID: 21148803 Free PMC article.
-
Thymic Determinants of γδ T Cell Differentiation.Trends Immunol. 2017 May;38(5):336-344. doi: 10.1016/j.it.2017.01.007. Epub 2017 Mar 9. Trends Immunol. 2017. PMID: 28285814 Review.
Cited by
-
Diversity of γδ T-cell antigens.Cell Mol Immunol. 2013 Jan;10(1):13-20. doi: 10.1038/cmi.2012.45. Epub 2012 Oct 22. Cell Mol Immunol. 2013. PMID: 23085946 Free PMC article. Review.
-
Development of γδ T Cells: Soldiers on the Front Lines of Immune Battles.Methods Mol Biol. 2023;2580:71-88. doi: 10.1007/978-1-0716-2740-2_4. Methods Mol Biol. 2023. PMID: 36374451
-
The transcriptional landscape of αβ T cell differentiation.Nat Immunol. 2013 Jun;14(6):619-32. doi: 10.1038/ni.2590. Epub 2013 May 5. Nat Immunol. 2013. PMID: 23644507 Free PMC article.
-
γδTCR-independent origin of neonatal γδ T cells prewired for IL-17 production.Curr Opin Immunol. 2019 Jun;58:60-67. doi: 10.1016/j.coi.2019.04.011. Epub 2019 May 23. Curr Opin Immunol. 2019. PMID: 31128446 Free PMC article. Review.
-
Developmental gene networks: a triathlon on the course to T cell identity.Nat Rev Immunol. 2014 Aug;14(8):529-45. doi: 10.1038/nri3702. Nat Rev Immunol. 2014. PMID: 25060579 Free PMC article. Review.
References
-
- Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. - PubMed
-
- Ferrick DA, et al. Differential production of interferon-γ and interleukin-4 in response to Th1-and Th2-stimulating pathogens by γδ T cells in vivo. Nature. 1995;373:255–257. - PubMed
-
- Stark MA, et al. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 2005;22:285–294. - PubMed
-
- Hayday A, Tigelaar R. Immunoregulation in the tissues by γδ T cells. Nature Rev Immunol. 2003;3:233–242. - PubMed
-
- Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nature Rev Immunol. 2007;7:479–485. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

