Short nucleosome repeats impose rotational modulations on chromatin fibre folding
- PMID: 22473209
- PMCID: PMC3364735
- DOI: 10.1038/emboj.2012.80
Short nucleosome repeats impose rotational modulations on chromatin fibre folding
Abstract
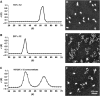
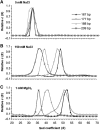
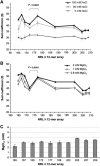
In eukaryotic cells, DNA is organized into arrays of repeated nucleosomes where the shorter nucleosome repeat length (NRL) types are associated with transcriptionally active chromatin. Here, we tested a hypothesis that systematic variations in the NRL influence nucleosome array folding into higher-order structures. For NRLs with fixed rotational settings, we observed a negative correlation between NRL and chromatin folding. Rotational variations within a range of longer NRLs (188 bp and above) typical of repressed chromatin in differentiated cells did not reveal any changes in chromatin folding. In sharp contrast, for the shorter NRL range of 165-177 bp, we observed a strong periodic dependence of chromatin folding upon the changes in linker DNA lengths, with the 172 bp repeat found in highly transcribed yeast chromatin imposing an unfolded state of the chromatin fibre that could be reversed by linker histone. Our results suggest that the NRL may direct chromatin higher-order structure into either a nucleosome position-dependent folding for short NRLs typical of transcribed genes or an architectural factor-dependent folding typical of longer NRLs prevailing in eukaryotic heterochromatin.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Carruthers LM, Bednar J, Woodcock CL, Hansen JC (1998) Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37: 14776–14787 - PubMed
-
- Carruthers LM, Schirf VR, Demeler B, Hansen JC (2000) Sedimentation velocity analysis of macromolecular assemblies. Methods Enzymol 321: 66–80 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

