Voltage-gated sodium channels at 60: structure, function and pathophysiology
- PMID: 22473783
- PMCID: PMC3424717
- DOI: 10.1113/jphysiol.2011.224204
Voltage-gated sodium channels at 60: structure, function and pathophysiology
Abstract
Voltage-gated sodium channels initiate action potentials in nerve, muscle and other excitable cells. The sodium current that initiates the nerve action potential was discovered by Hodgkin and Huxley using the voltage clamp technique in their landmark series of papers in The Journal of Physiology in 1952. They described sodium selectivity, voltage-dependent activation and fast inactivation, and they developed a quantitative model for action potential generation that has endured for many decades. This article gives an overview of the legacy that has evolved from their work, including development of conceptual models of sodium channel function, discovery of the sodium channel protein, analysis of its structure and function, determination of its structure at high resolution, definition of the mechanism and structural basis for drug block, and exploration of the role of the sodium channel as a target for disease mutations. Structural models for sodium selectivity and conductance, voltage-dependent activation, fast inactivation and drug block are discussed. A perspective for the future envisions new advances in understanding the structural basis for sodium channel function, the role of sodium channels in disease and the opportunity for discovery of novel therapeutics.
Figures


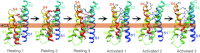
References
-
- Agnew WS, Moore AC, Levinson SR, Raftery MA. Identification of a large molecular weight peptide associated with a tetrodotoxin binding proteins from the electroplax of Electrophorus electricus. Biochem Biophys Res Commun. 1980;92:860–866. - PubMed
-
- Armstrong CM, Bezanilla F. Currents related to movement of the gating particles of the sodium channels. Nature. 1973;242:459–461. - PubMed
-
- Barchi RL. Protein components of the purified sodium channel from rat skeletal sarcolemma. J Neurochem. 1983;36:1377–1385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

