CONTRA: copy number analysis for targeted resequencing
- PMID: 22474122
- PMCID: PMC3348560
- DOI: 10.1093/bioinformatics/bts146
CONTRA: copy number analysis for targeted resequencing
Abstract
Motivation: In light of the increasing adoption of targeted resequencing (TR) as a cost-effective strategy to identify disease-causing variants, a robust method for copy number variation (CNV) analysis is needed to maximize the value of this promising technology.
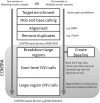
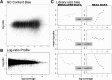
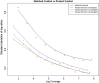
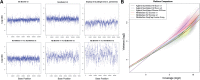
Results: We present a method for CNV detection for TR data, including whole-exome capture data. Our method calls copy number gains and losses for each target region based on normalized depth of coverage. Our key strategies include the use of base-level log-ratios to remove GC-content bias, correction for an imbalanced library size effect on log-ratios, and the estimation of log-ratio variations via binning and interpolation. Our methods are made available via CONTRA (COpy Number Targeted Resequencing Analysis), a software package that takes standard alignment formats (BAM/SAM) and outputs in variant call format (VCF4.0), for easy integration with other next-generation sequencing analysis packages. We assessed our methods using samples from seven different target enrichment assays, and evaluated our results using simulated data and real germline data with known CNV genotypes.
Figures

References
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995;57:289–300.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

