Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function
- PMID: 22474260
- PMCID: PMC3323877
- DOI: 10.1101/gad.186858.112
Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function
Abstract
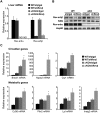
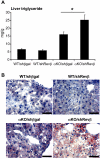
The nuclear receptor Rev-erbα regulates circadian rhythm and metabolism, but its effects are modest and it has been considered to be a secondary regulator of the cell-autonomous clock. Here we report that depletion of Rev-erbα together with closely related Rev-erbβ has dramatic effects on the cell-autonomous clock as well as hepatic lipid metabolism. Mouse embryonic fibroblasts were rendered arrhythmic by depletion of both Rev-erbs. In mouse livers, Rev-erbβ mRNA and protein levels oscillate with a diurnal pattern similar to that of Rev-erbα, and both Rev-erbs are recruited to a remarkably similar set of binding sites across the genome, enriched near metabolic genes. Depletion of both Rev-erbs in liver synergistically derepresses several metabolic genes as well as genes that control the positive limb of the molecular clock. Moreover, deficiency of both Rev-erbs causes marked hepatic steatosis, in contrast to relatively subtle changes upon loss of either subtype alone. These findings establish the two Rev-erbs as major regulators of both clock function and metabolism, displaying a level of subtype collaboration that is unusual among nuclear receptors but common among core clock proteins, protecting the organism from major perturbations in circadian and metabolic physiology.
Figures

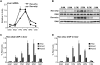


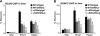
Comment in
-
REV-ERBs: more than the sum of the individual parts.Cell Metab. 2012 Jun 6;15(6):791-3. doi: 10.1016/j.cmet.2012.05.006. Cell Metab. 2012. PMID: 22682217
References
-
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR 2001. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron 30: 525–536 - PubMed
-
- Bonnelye E, Vanacker JM, Desbiens X, Begue A, Stehelin D, Laudet V 1994. Rev-erbβ, a new member of the nuclear receptor superfamily, is expressed in the nervous system during chicken development. Cell Growth Differ 5: 1357–1365 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
