Biotinylation of lysine method identifies acetylated histone H3 lysine 79 in Saccharomyces cerevisiae as a substrate for Sir2
- PMID: 22474337
- PMCID: PMC3341040
- DOI: 10.1073/pnas.1121471109
Biotinylation of lysine method identifies acetylated histone H3 lysine 79 in Saccharomyces cerevisiae as a substrate for Sir2
Abstract
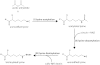
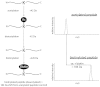
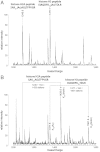
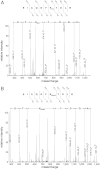
Although the biological roles of many members of the sirtuin family of lysine deacetylases have been well characterized, a broader understanding of their role in biology is limited by the challenges in identifying new substrates. We present here an in vitro method that combines biotinylation and mass spectrometry (MS) to identify substrates deacetylated by sirtuins. The method permits labeling of deacetylated residues with amine-reactive biotin on the ε-nitrogen of lysine. The biotin can be utilized to purify the substrate and identify the deacetylated lysine by MS. The biotinyl-lysine method was used to compare deacetylation of chemically acetylated histones by the yeast sirtuins, Sir2 and Hst2. Intriguingly, Sir2 preferentially deacetylates histone H3 lysine 79 as compared to Hst2. Although acetylation of K79 was not previously reported in Saccharomyces cerevisiae, we demonstrate that a minor population of this residue is indeed acetylated in vivo and show that Sir2, and not Hst2, regulates the acetylation state of H3 lysine 79. The in vitro biotinyl-lysine method combined with chemical acetylation made it possible to identify this previously unknown, low-abundance histone acetyl modification in vivo. This method has further potential to identify novel sirtuin deacetylation substrates in whole cell extracts, enabling large-scale screens for new deacetylase substrates.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. - PubMed
-
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. - PubMed
-
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

