Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA
- PMID: 22474350
- PMCID: PMC3341030
- DOI: 10.1073/pnas.1113532109
Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA
Abstract
B-DNA becomes unstable under superhelical stress and is able to adopt a wide range of alternative conformations including strand-separated DNA and Z-DNA. Localized sequence-dependent structural transitions are important for the regulation of biological processes such as DNA replication and transcription. To directly probe the effect of sequence on structural transitions driven by torque, we have measured the torsional response of a panel of DNA sequences using single molecule assays that employ nanosphere rotational probes to achieve high torque resolution. The responses of Z-forming d(pGpC)(n) sequences match our predictions based on a theoretical treatment of cooperative transitions in helical polymers. "Bubble" templates containing 50-100 bp mismatch regions show cooperative structural transitions similar to B-DNA, although less torque is required to disrupt strand-strand interactions. Our mechanical measurements, including direct characterization of the torsional rigidity of strand-separated DNA, establish a framework for quantitative predictions of the complex torsional response of arbitrary sequences in their biological context.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
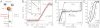
 (where l is the length of the DNA and Pt,eff is the effective twist persistence length) can be extracted from the torque-twist plot. DNA twist and rotor angular velocities were obtained from raw traces (insert) showing the cumulative angles of the magnets (orange trace) and RB (green trace) as a function of time. (C) Torque-twist plot showing complete conversion of B-DNA to L-DNA under negative torque for an N base pair long control DNA held under 3.2 pN tension. A faster twist ramp and proportionately larger bin sizes have been used for data in the plateau region. By saturating the transition, the extrapolated change in helicity per base pair Δθ0,L can be obtained. From the slope κL the twist persistence length
(where l is the length of the DNA and Pt,eff is the effective twist persistence length) can be extracted from the torque-twist plot. DNA twist and rotor angular velocities were obtained from raw traces (insert) showing the cumulative angles of the magnets (orange trace) and RB (green trace) as a function of time. (C) Torque-twist plot showing complete conversion of B-DNA to L-DNA under negative torque for an N base pair long control DNA held under 3.2 pN tension. A faster twist ramp and proportionately larger bin sizes have been used for data in the plateau region. By saturating the transition, the extrapolated change in helicity per base pair Δθ0,L can be obtained. From the slope κL the twist persistence length  of L-DNA can be calculated. (D) L-DNA helicity and elasticity vary with tension. Δθ0,L and effective twist persistence length Pt,eff,L of the L state are displayed ± SEM, based on linear fits to the data spanning −15 and -20 pNnm. Averaged traces are each based on three independent measurements.
of L-DNA can be calculated. (D) L-DNA helicity and elasticity vary with tension. Δθ0,L and effective twist persistence length Pt,eff,L of the L state are displayed ± SEM, based on linear fits to the data spanning −15 and -20 pNnm. Averaged traces are each based on three independent measurements.


Similar articles
-
Competition between B-Z and B-L transitions in a single DNA molecule: Computational studies.Phys Rev E. 2016 Feb;93(2):022411. doi: 10.1103/PhysRevE.93.022411. Epub 2016 Feb 19. Phys Rev E. 2016. PMID: 26986366
-
Theoretical analysis of competing conformational transitions in superhelical DNA.PLoS Comput Biol. 2012;8(4):e1002484. doi: 10.1371/journal.pcbi.1002484. Epub 2012 Apr 26. PLoS Comput Biol. 2012. PMID: 22570598 Free PMC article.
-
Sequence-dependent cost for Z-form shapes the torsion-driven B-Z transition via close interplay of Z-DNA and DNA bubble.Nucleic Acids Res. 2021 Apr 19;49(7):3651-3660. doi: 10.1093/nar/gkab153. Nucleic Acids Res. 2021. PMID: 33744929 Free PMC article.
-
Local supercoil-stabilized DNA structures.Crit Rev Biochem Mol Biol. 1991;26(2):151-226. doi: 10.3109/10409239109081126. Crit Rev Biochem Mol Biol. 1991. PMID: 1914495 Review.
-
Helix opening transitions in supercoiled DNA.Biochim Biophys Acta. 1992 May 7;1131(1):1-15. doi: 10.1016/0167-4781(92)90091-d. Biochim Biophys Acta. 1992. PMID: 1581350 Review. No abstract available.
Cited by
-
Probing the salt dependence of the torsional stiffness of DNA by multiplexed magnetic torque tweezers.Nucleic Acids Res. 2017 Jun 2;45(10):5920-5929. doi: 10.1093/nar/gkx280. Nucleic Acids Res. 2017. PMID: 28460037 Free PMC article.
-
Non-equilibrium structural dynamics of supercoiled DNA plasmids exhibits asymmetrical relaxation.Nucleic Acids Res. 2022 Mar 21;50(5):2754-2764. doi: 10.1093/nar/gkac101. Nucleic Acids Res. 2022. PMID: 35188541 Free PMC article.
-
Global force-torque phase diagram for the DNA double helix: structural transitions, triple points, and collapsed plectonemes.Phys Rev E Stat Nonlin Soft Matter Phys. 2013 Dec;88(6):062722. doi: 10.1103/PhysRevE.88.062722. Epub 2013 Dec 27. Phys Rev E Stat Nonlin Soft Matter Phys. 2013. PMID: 24483501 Free PMC article.
-
Multimodal Measurements of Single-Molecule Dynamics Using FluoRBT.Biophys J. 2018 Jan 23;114(2):278-282. doi: 10.1016/j.bpj.2017.11.017. Epub 2017 Dec 13. Biophys J. 2018. PMID: 29248150 Free PMC article.
-
Camera-based three-dimensional real-time particle tracking at kHz rates and Ångström accuracy.Nat Commun. 2015 Jan 7;6:5885. doi: 10.1038/ncomms6885. Nat Commun. 2015. PMID: 25565216 Free PMC article.
References
-
- Benham CJ. Energetics of the strand separation transition in superhelical DNA. J Mol Biol. 1992;225:835–847. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

