RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities
- PMID: 22474362
- PMCID: PMC3341018
- DOI: 10.1073/pnas.1119540109
RNA signatures allow rapid identification of pathogens and antibiotic susceptibilities
Abstract
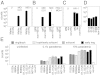
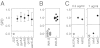
With rising rates of drug-resistant infections, there is a need for diagnostic methods that rapidly can detect the presence of pathogens and reveal their susceptibility to antibiotics. Here we propose an approach to diagnosing the presence and drug-susceptibility of infectious diseases based on direct detection of RNA from clinical samples. We demonstrate that species-specific RNA signatures can be used to identify a broad spectrum of infectious agents, including bacteria, viruses, yeast, and parasites. Moreover, we show that the behavior of a small set of bacterial transcripts after a brief antibiotic pulse can rapidly differentiate drug-susceptible and -resistant organisms and that these measurements can be made directly from clinical materials. Thus, transcriptional signatures could form the basis of a uniform diagnostic platform applicable across a broad range of infectious agents.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Park MM, Davis AL, Schluger NW, Cohen H, Rom WN. Outcome of MDR-TB patients, 1983-1993. Prolonged survival with appropriate therapy. Am J Respir Crit Care Med. 1996;153:317–324. - PubMed
-
- Ince J, McNally A. Development of rapid, automated diagnostics for infectious disease: Advances and challenges. Expert Rev Med Devices. 2009;6:641–651. - PubMed
-
- Cunningham J, Perkins M. Diagnostics for Tuberculosis: Global Demand and Market Potential. Geneva: WHO; 2006.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

