RNA ligase RtcB splices 3'-phosphate and 5'-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3')pp(5')G intermediates
- PMID: 22474365
- PMCID: PMC3341019
- DOI: 10.1073/pnas.1201207109
RNA ligase RtcB splices 3'-phosphate and 5'-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3')pp(5')G intermediates
Abstract
A cherished tenet of nucleic acid enzymology holds that synthesis of polynucleotide 3'-5' phosphodiesters proceeds via the attack of a 3'-OH on a high-energy 5' phosphoanhydride: either a nucleoside 5'-triphosphate in the case of RNA/DNA polymerases or an adenylylated intermediate A(5')pp(5')N--in the case of polynucleotide ligases. RtcB exemplifies a family of RNA ligases implicated in tRNA splicing and repair. Unlike classic ligases, RtcB seals broken RNAs with 3'-phosphate and 5'-OH ends. Here we show that RtcB executes a three-step ligation pathway entailing (i) reaction of His337 of the enzyme with GTP to form a covalent RtcB-(histidinyl-N)-GMP intermediate; (ii) transfer of guanylate to a polynucleotide 3'-phosphate to form a polynucleotide-(3')pp(5')G intermediate; and (iii) attack of a 5'-OH on the -N(3')pp(5')G end to form the splice junction. RtcB is structurally sui generis, and its chemical mechanism is unique. The wide distribution of RtcB proteins in bacteria, archaea, and metazoa raises the prospect of an alternative enzymology based on covalently activated 3' ends.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
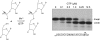

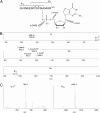


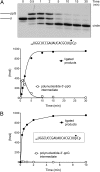
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

