HIV-1 antiretroviral drug therapy
- PMID: 22474613
- PMCID: PMC3312400
- DOI: 10.1101/cshperspect.a007161
HIV-1 antiretroviral drug therapy
Abstract
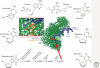
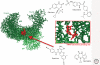
The most significant advance in the medical management of HIV-1 infection has been the treatment of patients with antiviral drugs, which can suppress HIV-1 replication to undetectable levels. The discovery of HIV-1 as the causative agent of AIDS together with an ever-increasing understanding of the virus replication cycle have been instrumental in this effort by providing researchers with the knowledge and tools required to prosecute drug discovery efforts focused on targeted inhibition with specific pharmacological agents. To date, an arsenal of 24 Food and Drug Administration (FDA)-approved drugs are available for treatment of HIV-1 infections. These drugs are distributed into six distinct classes based on their molecular mechanism and resistance profiles: (1) nucleoside-analog reverse transcriptase inhibitors (NNRTIs), (2) non-nucleoside reverse transcriptase inhibitors (NNRTIs), (3) integrase inhibitors, (4) protease inhibitors (PIs), (5) fusion inhibitors, and (6) coreceptor antagonists. In this article, we will review the basic principles of antiretroviral drug therapy, the mode of drug action, and the factors leading to treatment failure (i.e., drug resistance).
Figures





References
-
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA 1996. CC CKR5: A RANTES, MIP-1a, MIP-1b receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272: 1955–1958 - PubMed
-
- Antinori A, Zaccarelli M, Cingolani A, Forbici F, Rizzo MG, Trotta MP, Di Giambenedetto S, Narciso P, Ammassari A, Girardi E, et al. 2002. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS Res Hum Retroviruses 18: 835–838 - PubMed
-
- Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): Increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37: 15908–15917 - PubMed
-
- Autran B, Carcelain G, Li TS, Blanc C, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J 1997. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science 277: 112–116 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
