Quality-score refinement of SSU rRNA gene pyrosequencing differs across gene region for environmental samples
- PMID: 22476815
- PMCID: PMC3391548
- DOI: 10.1007/s00248-012-0043-9
Quality-score refinement of SSU rRNA gene pyrosequencing differs across gene region for environmental samples
Abstract
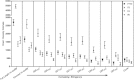
Due to potential sequencing errors in pyrosequencing data, species richness and diversity indices of microbial systems can be miscalculated. The "traditional" sequence refinement method is not sufficient to account for overestimations (e.g., length, primer errors, ambiguous nucleotides). Recent in silico and single-organism studies have revealed the importance of sequence quality scores in the estimation of ecological indices; however, this is the first study to compare quality-score stringencies across four regions of the SSU rRNA gene sequence (V1V2, V3, V4, and V6) with actual environmental samples compared directly to corresponding clone libraries produced from the same primer sets. The nucleic acid sequences determined via pyrosequencing were subjected to varying quality-score cutoffs that ranged from 25 to 32, and at each quality-score cutoff, either 10 or 15 % of the nucleotides were allowed to be below the cutoff. When species richness estimates were compared for the tested samples, the cutoff values of Q27(15%), Q30(10%), and Q32(15%) for V1V2, V4, and V6, respectively, estimated similar values as obtained with clone libraries and Sanger sequencing. The most stringent Q tested (Q32(10%)) was not enough to account for species richness inflation of the V3 region pyrosequence data. Results indicated that quality-score assessment greatly improved estimates of ecological indices for environmental samples (species richness and α-diversity) and that the effect of quality-score filtering was region-dependent.
Figures




References
-
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen ZT, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu PG, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

