Patterns of linkage disequilibrium and association mapping in diploid alfalfa (M. sativa L.)
- PMID: 22476875
- PMCID: PMC3397135
- DOI: 10.1007/s00122-012-1854-2
Patterns of linkage disequilibrium and association mapping in diploid alfalfa (M. sativa L.)
Abstract
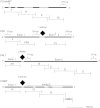
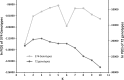
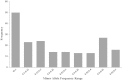
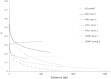
Association mapping enables the detection of marker-trait associations in unstructured populations by taking advantage of historical linkage disequilibrium (LD) that exists between a marker and the true causative polymorphism of the trait phenotype. Our first objective was to understand the pattern of LD decay in the diploid alfalfa genome. We used 89 highly polymorphic SSR loci in 374 unimproved diploid alfalfa (Medicago sativa L.) genotypes from 120 accessions to infer chromosome-wide patterns of LD. We also sequenced four lignin biosynthesis candidate genes (caffeoyl-CoA 3-O-methyltransferase (CCoAoMT), ferulate-5-hydroxylase (F5H), caffeic acid-O-methyltransferase (COMT), and phenylalanine amonialyase (PAL 1)) to identify single nucleotide polymorphisms (SNPs) and infer within gene estimates of LD. As the second objective of this study, we conducted association mapping for cell wall components and agronomic traits using the SSR markers and SNPs from the four candidate genes. We found very little LD among SSR markers implying limited value for genomewide association studies. In contrast, within gene LD decayed within 300 bp below an r (2) of 0.2 in three of four candidate genes. We identified one SSR and two highly significant SNPs associated with biomass yield. Based on our results, focusing association mapping on candidate gene sequences will be necessary until a dense set of genome-wide markers is available for alfalfa.
Figures


Similar articles
-
Identification of loci controlling forage yield and nutritive value in diploid alfalfa using GBS-GWAS.Theor Appl Genet. 2017 Feb;130(2):261-268. doi: 10.1007/s00122-016-2782-3. Epub 2016 Sep 23. Theor Appl Genet. 2017. PMID: 27662844
-
Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers.BMC Plant Biol. 2003 Dec 19;3:9. doi: 10.1186/1471-2229-3-9. BMC Plant Biol. 2003. PMID: 14683527 Free PMC article.
-
Inferring population structure and genetic diversity of broad range of wild diploid alfalfa (Medicago sativa L.) accessions using SSR markers.Theor Appl Genet. 2010 Aug;121(3):403-15. doi: 10.1007/s00122-010-1319-4. Epub 2010 Mar 30. Theor Appl Genet. 2010. PMID: 20352180
-
QTL mapping using high-throughput sequencing.Methods Mol Biol. 2015;1284:257-85. doi: 10.1007/978-1-4939-2444-8_13. Methods Mol Biol. 2015. PMID: 25757777 Review.
-
Methods for linkage disequilibrium mapping in crops.Trends Plant Sci. 2007 Feb;12(2):57-63. doi: 10.1016/j.tplants.2006.12.001. Epub 2007 Jan 16. Trends Plant Sci. 2007. PMID: 17224302 Review.
Cited by
-
Employing genome-wide SNP discovery and genotyping strategy to extrapolate the natural allelic diversity and domestication patterns in chickpea.Front Plant Sci. 2015 Mar 31;6:162. doi: 10.3389/fpls.2015.00162. eCollection 2015. Front Plant Sci. 2015. PMID: 25873920 Free PMC article.
-
Functionally relevant microsatellite markers from chickpea transcription factor genes for efficient genotyping applications and trait association mapping.DNA Res. 2013 Aug;20(4):355-74. doi: 10.1093/dnares/dst015. Epub 2013 Apr 29. DNA Res. 2013. PMID: 23633531 Free PMC article.
-
Genome-wide high-throughput SNP discovery and genotyping for understanding natural (functional) allelic diversity and domestication patterns in wild chickpea.Sci Rep. 2015 Jul 24;5:12468. doi: 10.1038/srep12468. Sci Rep. 2015. PMID: 26208313 Free PMC article.
-
ABC Transporter-Mediated Transport of Glutathione Conjugates Enhances Seed Yield and Quality in Chickpea.Plant Physiol. 2019 May;180(1):253-275. doi: 10.1104/pp.18.00934. Epub 2019 Feb 8. Plant Physiol. 2019. PMID: 30737266 Free PMC article.
-
Prospects for Trifolium Improvement Through Germplasm Characterisation and Pre-breeding in New Zealand and Beyond.Front Plant Sci. 2021 Jun 16;12:653191. doi: 10.3389/fpls.2021.653191. eCollection 2021. Front Plant Sci. 2021. PMID: 34220882 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

