The transcription factor ultraspiracle influences honey bee social behavior and behavior-related gene expression
- PMID: 22479195
- PMCID: PMC3315457
- DOI: 10.1371/journal.pgen.1002596
The transcription factor ultraspiracle influences honey bee social behavior and behavior-related gene expression
Abstract
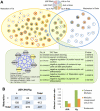
Behavior is among the most dynamic animal phenotypes, modulated by a variety of internal and external stimuli. Behavioral differences are associated with large-scale changes in gene expression, but little is known about how these changes are regulated. Here we show how a transcription factor (TF), ultraspiracle (usp; the insect homolog of the Retinoid X Receptor), working in complex transcriptional networks, can regulate behavioral plasticity and associated changes in gene expression. We first show that RNAi knockdown of USP in honey bee abdominal fat bodies delayed the transition from working in the hive (primarily "nursing" brood) to foraging outside. We then demonstrate through transcriptomics experiments that USP induced many maturation-related transcriptional changes in the fat bodies by mediating transcriptional responses to juvenile hormone. These maturation-related transcriptional responses to USP occurred without changes in USP's genomic binding sites, as revealed by ChIP-chip. Instead, behaviorally related gene expression is likely determined by combinatorial interactions between USP and other TFs whose cis-regulatory motifs were enriched at USP's binding sites. Many modules of JH- and maturation-related genes were co-regulated in both the fat body and brain, predicting that usp and cofactors influence shared transcriptional networks in both of these maturation-related tissues. Our findings demonstrate how "single gene effects" on behavioral plasticity can involve complex transcriptional networks, in both brain and peripheral tissues.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Osborne KA, Robichon A, Burgess E, Butland S, Shaw RA, et al. Natural behavior polymorphism due to a cGMP-dependent protein kinase of Drosophila. Science. 1997;277:834–836. - PubMed
-
- Haesler S, Rochefort C, Georgi B, Licznerski P, Osten P, et al. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. - DOI - PMC - PubMed
-
- Nelson CM, Ihle KE, Fondrk MK, Page RE, Amdam GV. The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol. 2007;5:e62. doi: 10.1371/journal.pbio.0050062. - DOI - PMC - PubMed
-
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

