Composition and evolution of the vertebrate and mammalian selenoproteomes
- PMID: 22479358
- PMCID: PMC3316567
- DOI: 10.1371/journal.pone.0033066
Composition and evolution of the vertebrate and mammalian selenoproteomes
Abstract
Background: Selenium is an essential trace element in mammals due to its presence in proteins in the form of selenocysteine (Sec). Human genome codes for 25 Sec-containing protein genes, and mouse and rat genomes for 24.
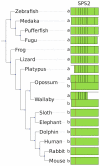
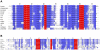
Methodology/principal findings: We characterized the selenoproteomes of 44 sequenced vertebrates by applying gene prediction and phylogenetic reconstruction methods, supplemented with the analyses of gene structures, alternative splicing isoforms, untranslated regions, SECIS elements, and pseudogenes. In total, we detected 45 selenoprotein subfamilies. 28 of them were found in mammals, and 41 in bony fishes. We define the ancestral vertebrate (28 proteins) and mammalian (25 proteins) selenoproteomes, and describe how they evolved along lineages through gene duplication (20 events), gene loss (10 events) and replacement of Sec with cysteine (12 events). We show that an intronless selenophosphate synthetase 2 gene evolved in early mammals and replaced functionally the original multiexon gene in placental mammals, whereas both genes remain in marsupials. Mammalian thioredoxin reductase 1 and thioredoxin-glutathione reductase evolved from an ancestral glutaredoxin-domain containing enzyme, still present in fish. Selenoprotein V and GPx6 evolved specifically in placental mammals from duplications of SelW and GPx3, respectively, and GPx6 lost Sec several times independently. Bony fishes were characterized by duplications of several selenoprotein families (GPx1, GPx3, GPx4, Dio3, MsrB1, SelJ, SelO, SelT, SelU1, and SelW2). Finally, we report identification of new isoforms for several selenoproteins and describe unusually conserved selenoprotein pseudogenes.
Conclusions/significance: This analysis represents the first comprehensive survey of the vertebrate and mammal selenoproteomes, and depicts their evolution along lineages. It also provides a wealth of information on these selenoproteins and their forms.
Conflict of interest statement
Figures







References
-
- Böck A, Forchhammer K, Heider J, Leinfelder W, Sawers G, et al. Selenocysteine: the 21st amino acid. Mol Microbiol. 1991;5:515–520. - PubMed
-
- Stadtman TC. Selenocysteine. Annu Rev Biochem. 1996;65:83–100. - PubMed
-
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, et al. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. - PubMed
-
- Low SC, Berry MJ. Knowing when to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci. 1996;21:203–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

