The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response
- PMID: 22479440
- PMCID: PMC3315573
- DOI: 10.1371/journal.pone.0033768
The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response
Abstract
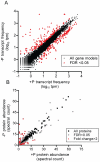
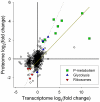
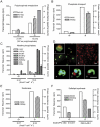
Phosphorus (P) is a critical driver of phytoplankton growth and ecosystem function in the ocean. Diatoms are an abundant class of marine phytoplankton that are responsible for significant amounts of primary production. With the control they exert on the oceanic carbon cycle, there have been a number of studies focused on how diatoms respond to limiting macro and micronutrients such as iron and nitrogen. However, diatom physiological responses to P deficiency are poorly understood. Here, we couple deep sequencing of transcript tags and quantitative proteomics to analyze the diatom Thalassiosira pseudonana grown under P-replete and P-deficient conditions. A total of 318 transcripts were differentially regulated with a false discovery rate of <0.05, and a total of 136 proteins were differentially abundant (p<0.05). Significant changes in the abundance of transcripts and proteins were observed and coordinated for multiple biochemical pathways, including glycolysis and translation. Patterns in transcript and protein abundance were also linked to physiological changes in cellular P distributions, and enzyme activities. These data demonstrate that diatom P deficiency results in changes in cellular P allocation through polyphosphate production, increased P transport, a switch to utilization of dissolved organic P through increased production of metalloenzymes, and a remodeling of the cell surface through production of sulfolipids. Together, these findings reveal that T. pseudonana has evolved a sophisticated response to P deficiency involving multiple biochemical strategies that are likely critical to its ability to respond to variations in environmental P availability.
Conflict of interest statement
Figures
References
-
- Dyhrman ST, Ammerman JW, Van Mooy BAS. Microbes and the Marine Phosphorus Cycle. Oceanography. 2007;20:110–116.
-
- Benitez-Nelson CR. The biogeochemical cycling of phosphorus in marine systems. Erth-Sci Rev. 2000;51:109–135.
-
- Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner B. Production and dissolution of biogenic silica in the ocean - revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cy. 1995;9:359–372.
-
- Lomas MW, Burke AL, Lomas DA, Bell DW, Shen C, et al. Sargasso Sea phosphorus biogeochemistry: an important role for dissolved organic phosphorus (DOP). Biogeosciences. 2010;7:695–710.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

