Cross-species analyses identify the BNIP-2 and Cdc42GAP homology (BCH) domain as a distinct functional subclass of the CRAL_TRIO/Sec14 superfamily
- PMID: 22479462
- PMCID: PMC3313917
- DOI: 10.1371/journal.pone.0033863
Cross-species analyses identify the BNIP-2 and Cdc42GAP homology (BCH) domain as a distinct functional subclass of the CRAL_TRIO/Sec14 superfamily
Abstract
The CRAL_TRIO protein domain, which is unique to the Sec14 protein superfamily, binds to a diverse set of small lipophilic ligands. Similar domains are found in a range of different proteins including neurofibromatosis type-1, a Ras GTPase-activating Protein (RasGAP) and Rho guanine nucleotide exchange factors (RhoGEFs). Proteins containing this structural protein domain exhibit a low sequence similarity and ligand specificity while maintaining an overall characteristic three-dimensional structure. We have previously demonstrated that the BNIP-2 and Cdc42GAP Homology (BCH) protein domain, which shares a low sequence homology with the CRAL_TRIO domain, can serve as a regulatory scaffold that binds to Rho, RhoGEFs and RhoGAPs to control various cell signalling processes. In this work, we investigate 175 BCH domain-containing proteins from a wide range of different organisms. A phylogenetic analysis with ~100 CRAL_TRIO and similar domains from eight representative species indicates a clear distinction of BCH-containing proteins as a novel subclass within the CRAL_TRIO/Sec14 superfamily. BCH-containing proteins contain a hallmark sequence motif R(R/K)h(R/K)(R/K)NL(R/K)xhhhhHPs ('h' is large and hydrophobic residue and 's' is small and weekly polar residue) and can be further subdivided into three unique subtypes associated with BNIP-2-N, macro- and RhoGAP-type protein domains. A previously unknown group of genes encoding 'BCH-only' domains is also identified in plants and arthropod species. Based on an analysis of their gene-structure and their protein domain context we hypothesize that BCH domain-containing genes evolved through gene duplication, intron insertions and domain swapping events. Furthermore, we explore the point of divergence between BCH and CRAL-TRIO proteins in relation to their ability to bind small GTPases, GAPs and GEFs and lipid ligands. Our study suggests a need for a more extensive analysis of previously uncharacterized BCH, 'BCH-like' and CRAL_TRIO-containing proteins and their significance in regulating signaling events involving small GTPases.
Conflict of interest statement
Figures



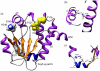
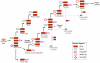
References
-
- Panagabko C, Morley S, Hernandez M, Cassolato P, Gordon H, et al. Ligand Specificity in the CRAL-TRIO Protein Family. Biochemistry. 2003;42:6467–6474. - PubMed
-
- Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

