Network-based functional enrichment
- PMID: 22479706
- PMCID: PMC3278830
- DOI: 10.1186/1471-2105-12-s13-s14
Network-based functional enrichment
Abstract
Background: Many methods have been developed to infer and reason about molecular interaction networks. These approaches often yield networks with hundreds or thousands of nodes and up to an order of magnitude more edges. It is often desirable to summarize the biological information in such networks. A very common approach is to use gene function enrichment analysis for this task. A major drawback of this method is that it ignores information about the edges in the network being analyzed, i.e., it treats the network simply as a set of genes. In this paper, we introduce a novel method for functional enrichment that explicitly takes network interactions into account.
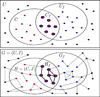
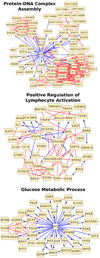
Results: Our approach naturally generalizes Fisher's exact test, a gene set-based technique. Given a function of interest, we compute the subgraph of the network induced by genes annotated to this function. We use the sequence of sizes of the connected components of this sub-network to estimate its connectivity. We estimate the statistical significance of the connectivity empirically by a permutation test. We present three applications of our method: i) determine which functions are enriched in a given network, ii) given a network and an interesting subnetwork of genes within that network, determine which functions are enriched in the sub-network, and iii) given two networks, determine the functions for which the connectivity improves when we merge the second network into the first. Through these applications, we show that our approach is a natural alternative to network clustering algorithms.
Conclusions: We presented a novel approach to functional enrichment that takes into account the pairwise relationships among genes annotated by a particular function. Each of the three applications discovers highly relevant functions. We used our methods to study biological data from three different organisms. Our results demonstrate the wide applicability of our methods. Our algorithms are implemented in C++ and are freely available under the GNU General Public License at our supplementary website. Additionally, all our input data andresults are available at http://bioinformatics.cs.vt.edu/~murali/supplements/2011-incob-nbe/.
Figures


References
-
- Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, Stark C, Ho Y, Botstein D, Andrews B, Boone C, Troyanskya OG, Ideker T, Dolinski K, Batada NN, Tyers M. Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. Journal of Biology. 2006;5(4):11+. doi: 10.1186/jbiol36. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

