A low-cost exon capture method suitable for large-scale screening of genetic deafness by the massively-parallel sequencing approach
- PMID: 22480152
- PMCID: PMC3378026
- DOI: 10.1089/gtmb.2011.0187
A low-cost exon capture method suitable for large-scale screening of genetic deafness by the massively-parallel sequencing approach
Abstract
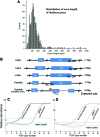
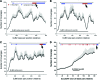
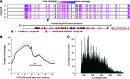
Current major barriers for using next-generation sequencing (NGS) technologies in genetic mutation screening on an epidemiological scale appear to be the high accuracy demanded by clinical applications and high per-sample cost. How to achieve high efficiency in enriching targeted disease genes while keeping a low cost/sample is a key technical hurdle to overcome. We validated a cDNA-probe-based approach for capturing exons of a group of genes known to cause deafness. Polymerase chain reaction amplicons were made from cDNA clones of the targeted genes and used as bait probes in hybridization for capturing human genomic DNA (gDNA) fragments. The cDNA library containing the clones of targeted genes provided a readily available, low-cost, and regenerable source for producing capture probes with standard molecular biology equipment. Captured gDNA fragments by our method were sequenced by the Illumina NGS platform. Results demonstrated that targeted exons captured by our approach achieved specificity, multiplexicity, uniformity, and depth of coverage suitable for accurate sequencing applications by the NGS systems. Reliable genotype calls for various homozygous and heterozygous mutations were achieved. The results were confirmed independently by conventional Sanger sequencing. The method validated here could be readily expanded to include all-known deafness genes for applications such as genetic hearing screening in newborns. The high coverage depth and cost benefits of the cDNA-probe-based exon capture approach may also facilitate widespread applications in clinical practices beyond screening mutations in deafness genes.
Figures
References
-
- Albert TJ. Molla MN. Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4:903–905. - PubMed
-
- Bao H. Guo H. Wang J, et al. MapView: visualization of short reads alignment on a desktop computer. Bioinformatics. 2009;25:1554–1555. - PubMed
-
- Chang EH. Van Camp G. Smith RJ. The role of connexins in human disease. Ear Hear. 2003;24:314–323. - PubMed
-
- Clemens CJ. Davis SA. Minimizing false-positives in universal newborn hearing screening: a simple solution. Pediatrics. 2001;107:E29. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

