The spliceosome: a flexible, reversible macromolecular machine
- PMID: 22480731
- PMCID: PMC3508674
- DOI: 10.1016/j.tibs.2012.02.009
The spliceosome: a flexible, reversible macromolecular machine
Abstract
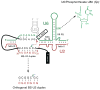
With more than a hundred individual RNA and protein parts and a highly dynamic assembly and disassembly pathway, the spliceosome is arguably the most complicated macromolecular machine in the eukaryotic cell. This complexity has made kinetic and mechanistic analysis of splicing incredibly challenging. Yet, recent technological advances are now providing tools for understanding this process in much greater detail. Ranging from genome-wide analyses of splicing and creation of an orthogonal spliceosome in vivo, to purification of active spliceosomes and observation of single molecules in vitro, such new experimental approaches are yielding significant insight into the inner workings of this remarkable machine. These experiments are rewriting the textbooks, with a new picture emerging of a dynamic, malleable machine heavily influenced by the identity of its pre-mRNA substrate.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


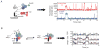

References
-
- Scherer S. Guide to the Human Genome. 1. Cold Spring Harbor; 2010.
-
- Wahl MC, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

