Complexity of RNA polymerase II elongation dynamics
- PMID: 22480952
- PMCID: PMC6310610
- DOI: 10.1016/j.bbagrm.2012.02.024
Complexity of RNA polymerase II elongation dynamics
Abstract
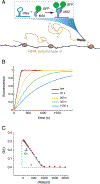
Transcription of protein-coding genes by RNA polymerase II can be regulated at multiple points during the process of RNA synthesis, including initiation, elongation, and termination. In vivo data suggests that elongating polymerases exhibit heterogeneity throughout the gene body, suggestive of changes in elongation rate and/or pausing. Here, we review evidence from a variety of different experimental approaches for understanding regulation of transcription elongation. We compare steady-state measurements of nascent RNA density and polymerase occupancy to time-resolved measurements and point out areas of disagreement. Finally, we discuss future avenues of investigation for understanding this critically important step in gene regulation. This article is part of a Special Issue entitled: Chromatin in time and space.
Published by Elsevier B.V.
Figures

References
-
- Sekimizu K, Kobayashi N, Mizuno D, Natori S, Purification of a factor from Ehrlich ascites tumor cells specifically stimulating RNA polymerase II, Biochemistry 15 (1976) 5064–5070. - PubMed
-
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y, Single-allele analysis of transcription kinetics in living mammalian cells, Nat. Methods 7 (2010) 631–633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

