Trypanosomatid comparative genomics: Contributions to the study of parasite biology and different parasitic diseases
- PMID: 22481868
- PMCID: PMC3313497
- DOI: 10.1590/s1415-47572012005000008
Trypanosomatid comparative genomics: Contributions to the study of parasite biology and different parasitic diseases
Abstract
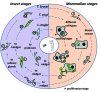
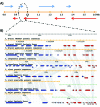
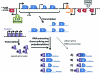
In 2005, draft sequences of the genomes of Trypanosoma brucei, Trypanosoma cruzi and Leishmania major, also known as the Tri-Tryp genomes, were published. These protozoan parasites are the causative agents of three distinct insect-borne diseases, namely sleeping sickness, Chagas disease and leishmaniasis, all with a worldwide distribution. Despite the large estimated evolutionary distance among them, a conserved core of ~6,200 trypanosomatid genes was found among the Tri-Tryp genomes. Extensive analysis of these genomic sequences has greatly increased our understanding of the biology of these parasites and their host-parasite interactions. In this article, we review the recent advances in the comparative genomics of these three species. This analysis also includes data on additional sequences derived from other trypanosmatid species, as well as recent data on gene expression and functional genomics. In addition to facilitating the identification of key parasite molecules that may provide a better understanding of these complex diseases, genome studies offer a rich source of new information that can be used to define potential new drug targets and vaccine candidates for controlling these parasitic infections.
Keywords: Leishmania major; RNAseq; Trypanosoma brucei; Trypanosoma cruzi; genome.
Figures

References
-
- Alves LR, Avila AR, Correa A, Holetz FB, Mansur FC, Manque PA, de Menezes JP, Buck GA, Krieger MA, Goldenberg S. Proteomic analysis reveals the dynamic association of proteins with translated mRNAs in Trypanosoma cruzi. Gene. 2010;452:72–78. - PubMed
-
- Araujo PR, Burle-Caldas GA, Silva-Pereira RA, Bartholomeu DC, DaRocha WD, Teixeira SM. Development of a dual reporter system to identify regulatory cis-acting elements in untranslated regions of Trypanosoma cruzi mRNAs. Parasitol Int. 2011;60:161–169. - PubMed
LinkOut - more resources
Full Text Sources
